Succession of sulfur-oxidizing bacteria in the microbial community on corroding concrete in sewer systems
- PMID: 17142362
- PMCID: PMC1800771
- DOI: 10.1128/AEM.02054-06
Succession of sulfur-oxidizing bacteria in the microbial community on corroding concrete in sewer systems
Abstract
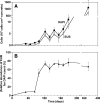
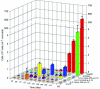
Microbially induced concrete corrosion (MICC) in sewer systems has been a serious problem for a long time. A better understanding of the succession of microbial community members responsible for the production of sulfuric acid is essential for the efficient control of MICC. In this study, the succession of sulfur-oxidizing bacteria (SOB) in the bacterial community on corroding concrete in a sewer system in situ was investigated over 1 year by culture-independent 16S rRNA gene-based molecular techniques. Results revealed that at least six phylotypes of SOB species were involved in the MICC process, and the predominant SOB species shifted in the following order: Thiothrix sp., Thiobacillus plumbophilus, Thiomonas intermedia, Halothiobacillus neapolitanus, Acidiphilium acidophilum, and Acidithiobacillus thiooxidans. A. thiooxidans, a hyperacidophilic SOB, was the most dominant (accounting for 70% of EUB338-mixed probe-hybridized cells) in the heavily corroded concrete after 1 year. This succession of SOB species could be dependent on the pH of the concrete surface as well as on trophic properties (e.g., autotrophic or mixotrophic) and on the ability of the SOB to utilize different sulfur compounds (e.g., H2S, S0, and S2O3(2-)). In addition, diverse heterotrophic bacterial species (e.g., halo-tolerant, neutrophilic, and acidophilic bacteria) were associated with these SOB. The microbial succession of these microorganisms was involved in the colonization of the concrete and the production of sulfuric acid. Furthermore, the vertical distribution of microbial community members revealed that A. thiooxidans was the most dominant throughout the heavily corroded concrete (gypsum) layer and that A. thiooxidans was most abundant at the highest surface (1.5-mm) layer and decreased logarithmically with depth because of oxygen and H2S transport limitations. This suggested that the production of sulfuric acid by A. thiooxidans occurred mainly on the concrete surface and the sulfuric acid produced penetrated through the corroded concrete layer and reacted with the sound concrete below.
Figures



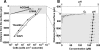
Similar articles
-
Characterization of sulfur oxidizing bacteria related to biogenic sulfuric acid corrosion in sludge digesters.BMC Microbiol. 2016 Jul 18;16(1):153. doi: 10.1186/s12866-016-0767-7. BMC Microbiol. 2016. PMID: 27430211 Free PMC article.
-
Microbial community structures and in situ sulfate-reducing and sulfur-oxidizing activities in biofilms developed on mortar specimens in a corroded sewer system.Water Res. 2009 Oct;43(18):4729-39. doi: 10.1016/j.watres.2009.07.035. Epub 2009 Aug 6. Water Res. 2009. PMID: 19709714
-
Revealing biogenic sulfuric acid corrosion in sludge digesters: detection of sulfur-oxidizing bacteria within full-scale digesters.Water Sci Technol. 2014;70(8):1405-11. doi: 10.2166/wst.2014.371. Water Sci Technol. 2014. PMID: 25353947
-
The Ecology of Acidophilic Microorganisms in the Corroding Concrete Sewer Environment.Front Microbiol. 2017 Apr 20;8:683. doi: 10.3389/fmicb.2017.00683. eCollection 2017. Front Microbiol. 2017. PMID: 28473816 Free PMC article. Review.
-
Acidithiobacillus thiooxidans and its potential application.Appl Microbiol Biotechnol. 2019 Oct;103(19):7819-7833. doi: 10.1007/s00253-019-10098-5. Epub 2019 Aug 28. Appl Microbiol Biotechnol. 2019. PMID: 31463545 Review.
Cited by
-
Biomineralization To Prevent Microbially Induced Corrosion on Concrete for Sustainable Marine Infrastructure.Environ Sci Technol. 2024 Jan 9;58(1):522-533. doi: 10.1021/acs.est.3c04680. Epub 2023 Dec 5. Environ Sci Technol. 2024. PMID: 38052449 Free PMC article.
-
Characterization of sulfur oxidizing bacteria related to biogenic sulfuric acid corrosion in sludge digesters.BMC Microbiol. 2016 Jul 18;16(1):153. doi: 10.1186/s12866-016-0767-7. BMC Microbiol. 2016. PMID: 27430211 Free PMC article.
-
Effect of Pseudomonas aeruginosa on corrosion of X65 pipeline steel.Heliyon. 2022 Dec 24;8(12):e12588. doi: 10.1016/j.heliyon.2022.e12588. eCollection 2022 Dec. Heliyon. 2022. PMID: 36643323 Free PMC article.
-
Influence of Dissolved-Aluminum Concentration on Sulfur-Oxidizing Bacterial Activity in the Biodeterioration of Concrete.Appl Environ Microbiol. 2019 Jul 18;85(15):e00302-19. doi: 10.1128/AEM.00302-19. Print 2019 Aug 1. Appl Environ Microbiol. 2019. PMID: 31126946 Free PMC article.
-
Identification of a novel acetate-utilizing bacterium belonging to Synergistes group 4 in anaerobic digester sludge.ISME J. 2011 Dec;5(12):1844-56. doi: 10.1038/ismej.2011.59. Epub 2011 May 12. ISME J. 2011. PMID: 21562600 Free PMC article.
References
-
- Amann, R. I. 1995. In situ identification of microorganisms by whole cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkerman, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
-
- Bond, P. L., and J. F. Banfield. 2001. Designed and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb. Ecol. 41:149-161. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

