Transcriptomic analysis reveals a bifurcated terephthalate degradation pathway in Rhodococcus sp. strain RHA1
- PMID: 17142403
- PMCID: PMC1855752
- DOI: 10.1128/JB.01322-06
Transcriptomic analysis reveals a bifurcated terephthalate degradation pathway in Rhodococcus sp. strain RHA1
Abstract
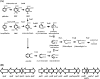
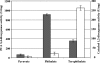
Phthalate isomers and their esters are important pollutants whose biodegradation is not well understood. Rhodococcus sp. strain RHA1 is notable for its ability to degrade a wide range of aromatic compounds. RHA1 was previously shown to degrade phthalate (PTH) and to have genes putatively encoding terephthalate (TPA) degradation. Transcriptomic analysis of 8,213 genes indicated that 150 were up-regulated during growth on PTH and that 521 were up-regulated during growth on TPA. Distinct ring cleavage dioxygenase systems were differentially expressed during growth on PTH and TPA. Genes encoding the protocatechuate (PCA) pathway were induced on both substrates, while genes encoding the catechol branch of the PCA pathway were additionally induced only on TPA. Accordingly, protocatechuate-3,4-dioxygenase activity was induced in cells grown on both substrates, while catechol-1,2-dioxygenase activity was induced only in cells grown on TPA. Knockout analysis indicated that pcaL, encoding 3-oxoadipate enol-lactone hydrolase and 4-carboxymuconolactone decarboxylase, was required for growth on both substrates but that pcaB, encoding beta-carboxy-cis,cis-muconate lactonizing enzyme, was required for growth on PTH only. These results indicate that PTH is degraded solely via the PCA pathway, whereas TPA is degraded via a bifurcated pathway that additionally includes the catechol branch of the PCA pathway.
Figures
Similar articles
-
Involvement of a novel ABC transporter and monoalkyl phthalate ester hydrolase in phthalate ester catabolism by Rhodococcus jostii RHA1.Appl Environ Microbiol. 2010 Mar;76(5):1516-23. doi: 10.1128/AEM.02621-09. Epub 2009 Dec 28. Appl Environ Microbiol. 2010. PMID: 20038686 Free PMC article.
-
Molecular and biochemical analysis of phthalate and terephthalate degradation by Rhodococcus sp. strain DK17.FEMS Microbiol Lett. 2005 Nov 15;252(2):207-13. doi: 10.1016/j.femsle.2005.08.045. Epub 2005 Sep 12. FEMS Microbiol Lett. 2005. PMID: 16181748
-
Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity.J Bacteriol. 1998 Mar;180(5):1072-81. doi: 10.1128/JB.180.5.1072-1081.1998. J Bacteriol. 1998. PMID: 9495744 Free PMC article.
-
Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: redundancies and convergence.J Bacteriol. 2005 Jun;187(12):4050-63. doi: 10.1128/JB.187.12.4050-4063.2005. J Bacteriol. 2005. PMID: 15937168 Free PMC article.
-
In vitro reconstitution of the catabolic reactions catalyzed by PcaHG, PcaB, and PcaL: the protocatechuate branch of the β-ketoadipate pathway in Rhodococcus jostii RHA1.Biosci Biotechnol Biochem. 2015;79(5):830-5. doi: 10.1080/09168451.2014.993915. Epub 2015 Jan 3. Biosci Biotechnol Biochem. 2015. PMID: 25558786
Cited by
-
Transcriptional regulation of the terephthalate catabolism operon in Comamonas sp. strain E6.Appl Environ Microbiol. 2010 Sep;76(18):6047-55. doi: 10.1128/AEM.00742-10. Epub 2010 Jul 23. Appl Environ Microbiol. 2010. PMID: 20656871 Free PMC article.
-
A Multi-Streamline Approach for Upcycling PET into a Biodiesel and Asphalt Modifier.Polymers (Basel). 2024 Mar 13;16(6):796. doi: 10.3390/polym16060796. Polymers (Basel). 2024. PMID: 38543401 Free PMC article.
-
Transcriptomic Analysis of Rhodococcus opacus R7 Grown on o-Xylene by RNA-Seq.Front Microbiol. 2020 Aug 12;11:1808. doi: 10.3389/fmicb.2020.01808. eCollection 2020. Front Microbiol. 2020. PMID: 32903390 Free PMC article.
-
Involvement of a novel ABC transporter and monoalkyl phthalate ester hydrolase in phthalate ester catabolism by Rhodococcus jostii RHA1.Appl Environ Microbiol. 2010 Mar;76(5):1516-23. doi: 10.1128/AEM.02621-09. Epub 2009 Dec 28. Appl Environ Microbiol. 2010. PMID: 20038686 Free PMC article.
-
Structural Insights into Dihydroxylation of Terephthalate, a Product of Polyethylene Terephthalate Degradation.J Bacteriol. 2022 Mar 15;204(3):e0054321. doi: 10.1128/JB.00543-21. Epub 2022 Jan 10. J Bacteriol. 2022. PMID: 35007143 Free PMC article.
References
-
- Batie, C. J., E. LaHaie, and D. P. Ballou. 1987. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J. Biol. Chem. 262:1510-1518. - PubMed
-
- Cain, R. B. 1966. Utilization of anthranilic and nitrobenzoic acids by Nocardia opaca and a flavobacterium. J. Gen. Microbiol. 42:219-235. - PubMed
-
- Cartwright, C. D., S. A. Owen, I. P. Thompson, and R. G. Burns. 2000. Biodegradation of diethyl phthalate in soil by a novel pathway. FEMS Microbiol. Lett. 186:27-34. - PubMed
-
- Chang, B. V., C. S. Liao, and S. Y. Yuan. 2005. Anaerobic degradation of diethyl phthalate, di-n-butyl phthalate, and di-(2-ethylhexyl) phthalate from river sediment in Taiwan. Chemosphere 58:1601-1607. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

