Conservation of the salt overly sensitive pathway in rice
- PMID: 17142477
- PMCID: PMC1803719
- DOI: 10.1104/pp.106.092635
Conservation of the salt overly sensitive pathway in rice
Abstract
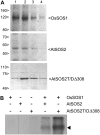
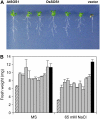
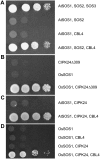
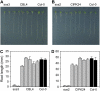
The salt tolerance of rice (Oryza sativa) correlates with the ability to exclude Na+ from the shoot and to maintain a low cellular Na+/K+ ratio. We have identified a rice plasma membrane Na+/H+ exchanger that, on the basis of genetic and biochemical criteria, is the functional homolog of the Arabidopsis (Arabidopsis thaliana) salt overly sensitive 1 (SOS1) protein. The rice transporter, denoted by OsSOS1, demonstrated a capacity for Na+/H+ exchange in plasma membrane vesicles of yeast (Saccharomyces cerevisiae) cells and reduced their net cellular Na+ content. The Arabidopsis protein kinase complex SOS2/SOS3, which positively controls the activity of AtSOS1, phosphorylated OsSOS1 and stimulated its activity in vivo and in vitro. Moreover, OsSOS1 suppressed the salt sensitivity of a sos1-1 mutant of Arabidopsis. These results represent the first molecular and biochemical characterization of a Na+ efflux protein from monocots. Putative rice homologs of the Arabidopsis protein kinase SOS2 and its Ca2+-dependent activator SOS3 were identified also. OsCIPK24 and OsCBL4 acted coordinately to activate OsSOS1 in yeast cells and they could be exchanged with their Arabidopsis counterpart to form heterologous protein kinase modules that activated both OsSOS1 and AtSOS1 and suppressed the salt sensitivity of sos2 and sos3 mutants of Arabidopsis. These results demonstrate that the SOS salt tolerance pathway operates in cereals and evidences a high degree of structural conservation among the SOS proteins from dicots and monocots.
Figures

Similar articles
-
Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana.Plant Cell. 2004 Feb;16(2):435-49. doi: 10.1105/tpc.019174. Epub 2004 Jan 23. Plant Cell. 2004. PMID: 14742879 Free PMC article.
-
Comparing Essentiality of SOS1-Mediated Na+ Exclusion in Salinity Tolerance between Cultivated and Wild Rice Species.Int J Mol Sci. 2022 Aug 31;23(17):9900. doi: 10.3390/ijms23179900. Int J Mol Sci. 2022. PMID: 36077294 Free PMC article.
-
The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance.J Exp Bot. 2020 Mar 25;71(6):1801-1814. doi: 10.1093/jxb/erz549. J Exp Bot. 2020. PMID: 31858132 Free PMC article.
-
Ion homeostasis during salt stress in plants.Curr Opin Cell Biol. 2001 Aug;13(4):399-404. doi: 10.1016/s0955-0674(00)00227-1. Curr Opin Cell Biol. 2001. PMID: 11454443 Review.
-
A Salt Overly Sensitive Pathway Member from Brassica juncea BjSOS3 Can Functionally Complement ΔAtsos3 in Arabidopsis.Curr Genomics. 2018 Jan;19(1):60-69. doi: 10.2174/1389202918666170228133621. Curr Genomics. 2018. PMID: 29491733 Free PMC article. Review.
Cited by
-
Quantitative expression analysis of TaSOS1 and TaSOS4 genes in cultivated and wild wheat plants under salt stress.Mol Biotechnol. 2013 Feb;53(2):189-97. doi: 10.1007/s12033-012-9513-z. Mol Biotechnol. 2013. PMID: 22367644
-
Molecular cloning and functional characterization of a novel apple MdCIPK6L gene reveals its involvement in multiple abiotic stress tolerance in transgenic plants.Plant Mol Biol. 2012 May;79(1-2):123-35. doi: 10.1007/s11103-012-9899-9. Epub 2012 Mar 1. Plant Mol Biol. 2012. PMID: 22382993
-
Comparative analysis of stress-induced calcium signals in the crop species barley and the model plant Arabidopsis thaliana.BMC Plant Biol. 2022 Sep 17;22(1):447. doi: 10.1186/s12870-022-03820-5. BMC Plant Biol. 2022. PMID: 36114461 Free PMC article.
-
Comparative proteomic analysis of early salt stress-responsive proteins in roots of SnRK2 transgenic rice.Proteome Sci. 2012 Mar 31;10:25. doi: 10.1186/1477-5956-10-25. Proteome Sci. 2012. PMID: 22462395 Free PMC article.
-
Seed Halopriming Improves Salinity Tolerance of Some Rice Cultivars During Seedling Stage.Bot Stud. 2022 Jul 25;63(1):24. doi: 10.1186/s40529-022-00354-9. Bot Stud. 2022. PMID: 35877013 Free PMC article.
References
-
- Bañuelos MA, Sychrová H, Bleykasten-Grosshans C, Souciet JL, Potier S (1998) The Nha1 antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology 144 2749–2758 - PubMed
-
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

