The strength of the HIV-1 3' splice sites affects Rev function
- PMID: 17144911
- PMCID: PMC1697824
- DOI: 10.1186/1742-4690-3-89
The strength of the HIV-1 3' splice sites affects Rev function
Abstract
Background: The HIV-1 Rev protein is a key component in the early to late switch in HIV-1 splicing from early intronless (e.g. tat, rev) to late intron-containing Rev-dependent (e.g. gag, vif, env) transcripts. Previous results suggested that cis-acting sequences and inefficient 5' and 3' splice sites are a prerequisite for Rev function. However, we and other groups have shown that two of the HIV-1 5' splice sites, D1 and D4, are efficiently used in vitro and in vivo. Here, we focus on the efficiency of the HIV-1 3' splice sites taking into consideration to what extent their intrinsic efficiencies are modulated by their downstream cis-acting exonic sequences. Furthermore, we delineate their role in RNA stabilization and Rev function.
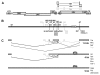
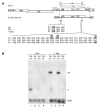
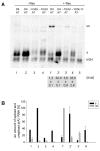
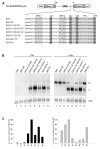
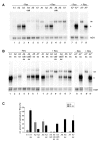
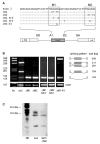
Results: In the presence of an efficient upstream 5' splice site the integrity of the 3' splice site is not essential for Rev function whereas an efficient 3' splice site impairs Rev function. The detrimental effect of a strong 3' splice site on the amount of Rev-dependent intron-containing HIV-1 glycoprotein coding (env) mRNA is not compensatable by weakening the strength of the upstream 5' splice site. Swapping the HIV-1 3' splice sites in an RRE-containing minigene, we found a 3' splice site usage which was variably dependent on the presence of the usual downstream exonic sequence. The most evident activation of 3' splice site usage by its usual downstream exonic sequence was observed for 3' splice site A1 which was turned from an intrinsic very weak 3' splice site into the most active 3' splice site, even abolishing Rev activity. Performing pull-down experiments with nuclear extracts of HeLa cells we identified a novel ASF/SF2-dependent exonic splicing enhancer (ESE) within HIV-1 exon 2 consisting of a heptameric sequence motif occurring twice (M1 and M2) within this short non-coding leader exon. Single point mutation of M1 within an infectious molecular clone is detrimental for HIV-1 exon 2 recognition without affecting Rev-dependent vif expression.
Conclusion: Under the conditions of our assay, the rate limiting step of retroviral splicing, competing with Rev function, seems to be exclusively determined by the functional strength of the 3' splice site. The bipartite ASF/SF2-dependent ESE within HIV-1 exon 2 supports cross-talk between splice site pairs across exon 2 (exon definition) which is incompatible with processing of the intron-containing vif mRNA. We propose that Rev mediates a switch from exon to intron definition necessary for the expression of all intron-containing mRNAs.
Figures




Similar articles
-
Synergistic stimulation of HIV-1 rev-dependent export of unspliced mRNA to the cytoplasm by hnRNP A1.J Mol Biol. 1999 Feb 5;285(5):1951-64. doi: 10.1006/jmbi.1998.2473. J Mol Biol. 1999. PMID: 9925777
-
Biological importance and cooperativity of HIV-1 regulatory gene splice acceptors.Virology. 1994 Jul;202(1):264-71. doi: 10.1006/viro.1994.1342. Virology. 1994. PMID: 8009838
-
Differential effects of intronic and exonic locations of the human immunodeficiency virus type 1 Rev-responsive element.Virology. 1996 May 15;219(2):423-31. doi: 10.1006/viro.1996.0268. Virology. 1996. PMID: 8638408
-
Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication.Adv Virus Res. 2009;74:1-40. doi: 10.1016/S0065-3527(09)74001-1. Adv Virus Res. 2009. PMID: 19698894 Review.
-
A role for Rev in the association of HIV-1 gag mRNA with cytoskeletal beta-actin and viral protein expression.Biochimie. 1996;78(11-12):1075-80. doi: 10.1016/s0300-9084(97)86732-6. Biochimie. 1996. PMID: 9150887 Review.
Cited by
-
Identification of a heterogeneous nuclear ribonucleoprotein-recognition region in the HIV Rev protein.J Biol Chem. 2009 Nov 27;284(48):33384-91. doi: 10.1074/jbc.M109.021659. Epub 2009 Oct 5. J Biol Chem. 2009. PMID: 19808671 Free PMC article.
-
Global synonymous mutagenesis identifies cis-acting RNA elements that regulate HIV-1 splicing and replication.PLoS Pathog. 2018 Jan 29;14(1):e1006824. doi: 10.1371/journal.ppat.1006824. eCollection 2018 Jan. PLoS Pathog. 2018. PMID: 29377940 Free PMC article.
-
Negative and positive mRNA splicing elements act competitively to regulate human immunodeficiency virus type 1 vif gene expression.J Virol. 2008 Apr;82(8):3921-31. doi: 10.1128/JVI.01558-07. Epub 2008 Feb 13. J Virol. 2008. PMID: 18272582 Free PMC article.
-
HIV-1: To Splice or Not to Splice, That Is the Question.Viruses. 2021 Jan 26;13(2):181. doi: 10.3390/v13020181. Viruses. 2021. PMID: 33530363 Free PMC article. Review.
-
The influenza A virus spliced messenger RNA M mRNA3 is not required for viral replication in tissue culture.J Gen Virol. 2008 Dec;89(Pt 12):3097-3101. doi: 10.1099/vir.0.2008/004739-0. J Gen Virol. 2008. PMID: 19008398 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

