Kinetic studies of HIV-1 and HIV-2 envelope glycoprotein-mediated fusion
- PMID: 17144914
- PMCID: PMC1693918
- DOI: 10.1186/1742-4690-3-90
Kinetic studies of HIV-1 and HIV-2 envelope glycoprotein-mediated fusion
Abstract
Background: HIV envelope glycoprotein (Env)-mediated fusion is driven by the concerted coalescence of the HIV gp41 N-helical and C-helical regions, which results in the formation of 6 helix bundles. Kinetics of HIV Env-mediated fusion is an important determinant of sensitivity to entry inhibitors and antibodies. However, the parameters that govern the HIV Env fusion cascade have yet to be fully elucidated. We address this issue by comparing the kinetics HIV-1IIIB Env with those mediated by HIV-2 from two strains with different affinities for CD4 and CXCR4.
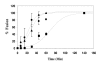
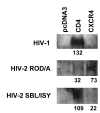
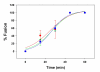
Results: HIV-1 and HIV-2 Env-mediated cell fusion occurred with half times of about 60 and 30 min, respectively. Binding experiments of soluble HIV gp120 proteins to CD4 and co-receptor did not correlate with the differences in kinetics of fusion mediated by the three different HIV Envs. However, escape from inhibition by reagents that block gp120-CD4 binding, CD4-induced CXCR4 binding and 6-helix bundle formation, respectively, indicated large difference between HIV-1 and HIV-2 envelope glycoproteins in their CD4-induced rates of engagement with CXCR4.
Conclusion: The HIV-2 Env proteins studied here exhibited a significantly reduced window of time between the engagement of gp120 with CD4 and exposure of the CXCR4 binding site on gp120 as compared with HIV-1IIIB Env. The efficiency with which HIV-2 Env undergoes this CD4-induced conformational change is the major cause of the relatively rapid rate of HIV-2 Env mediated-fusion.
Figures

Similar articles
-
HIV-1 gp41 six-helix bundle formation occurs rapidly after the engagement of gp120 by CXCR4 in the HIV-1 Env-mediated fusion process.Biochemistry. 2001 Oct 16;40(41):12231-6. doi: 10.1021/bi0155596. Biochemistry. 2001. PMID: 11591141
-
HIV-1 gp41 Residues Modulate CD4-Induced Conformational Changes in the Envelope Glycoprotein and Evolution of a Relaxed Conformation of gp120.J Virol. 2018 Jul 31;92(16):e00583-18. doi: 10.1128/JVI.00583-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29875245 Free PMC article.
-
Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics.Proc Natl Acad Sci U S A. 2002 Dec 10;99(25):16249-54. doi: 10.1073/pnas.252469399. Epub 2002 Nov 20. Proc Natl Acad Sci U S A. 2002. PMID: 12444251 Free PMC article.
-
CD4 activation of HIV fusion.Int J Cell Cloning. 1992 Nov;10(6):323-32. doi: 10.1002/stem.5530100603. Int J Cell Cloning. 1992. PMID: 1281202 Review.
-
The HIV Env-mediated fusion reaction.Biochim Biophys Acta. 2003 Jul 11;1614(1):36-50. doi: 10.1016/s0005-2736(03)00161-5. Biochim Biophys Acta. 2003. PMID: 12873764 Review.
Cited by
-
Distinct mechanisms regulate exposure of neutralizing epitopes in the V2 and V3 loops of HIV-1 envelope.J Virol. 2014 Nov;88(21):12853-65. doi: 10.1128/JVI.02125-14. Epub 2014 Aug 27. J Virol. 2014. PMID: 25165106 Free PMC article.
-
An antibody directed against the fusion peptide of Junin virus envelope glycoprotein GPC inhibits pH-induced membrane fusion.J Virol. 2010 Jun;84(12):6119-29. doi: 10.1128/JVI.02700-09. Epub 2010 Apr 14. J Virol. 2010. PMID: 20392854 Free PMC article.
-
Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme.Crit Rev Biochem Mol Biol. 2008 May-Jun;43(3):189-219. doi: 10.1080/10409230802058320. Crit Rev Biochem Mol Biol. 2008. PMID: 18568847 Free PMC article. Review.
-
Different infectivity of HIV-1 strains is linked to number of envelope trimers required for entry.PLoS Pathog. 2015 Jan 8;11(1):e1004595. doi: 10.1371/journal.ppat.1004595. eCollection 2015 Jan. PLoS Pathog. 2015. PMID: 25569556 Free PMC article.
-
The six-helix bundle of human immunodeficiency virus Env controls pore formation and enlargement and is initiated at residues proximal to the hairpin turn.J Virol. 2009 Oct;83(19):10048-57. doi: 10.1128/JVI.00316-09. Epub 2009 Jul 22. J Virol. 2009. PMID: 19625396 Free PMC article.
References
-
- Reeves JD, Doms RW. Human immunodeficiency virus type 2. J Gen Virol. 2002;83:1253–1265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

