MEKK4 signaling regulates filamin expression and neuronal migration
- PMID: 17145501
- PMCID: PMC1876745
- DOI: 10.1016/j.neuron.2006.10.024
MEKK4 signaling regulates filamin expression and neuronal migration
Abstract
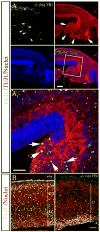
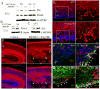
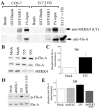
Periventricular heterotopia (PVH) is a congenital malformation of human cerebral cortex frequently associated with Filamin-A (FLN-A) mutations but the pathogenetic mechanisms remain unclear. Here, we show that the MEKK4 (MAP3K4) pathway is involved in Fln-A regulation and PVH formation. MEKK4(-/-) mice developed PVH associated with breaches in the neuroependymal lining which were largely comprised of neurons that failed to reach the cortical plate. RNA interference (RNAi) targeting MEKK4 also impaired neuronal migration. Expression of Fln was elevated in MEKK4(-/-) forebrain, most notably near sites of failed neuronal migration. Importantly, recombinant MKK4 protein precipitated a complex containing MEKK4 and Fln-A, and MKK4 mediated signaling between MEKK4 and Fln-A, suggesting that MKK4 may bridge these molecules during development. Finally, we showed that wild-type FLN-A overexpression inhibited neuronal migration. Collectively, our results demonstrate a link between MEKK4 and Fln-A that impacts neuronal migration initiation and provides insight into the pathogenesis of human PVH.
Figures




Similar articles
-
Trouble making the first move: interpreting arrested neuronal migration in the cerebral cortex.Trends Neurosci. 2008 Feb;31(2):54-61. doi: 10.1016/j.tins.2007.11.009. Epub 2008 Jan 16. Trends Neurosci. 2008. PMID: 18201775 Review.
-
Disruption of neural progenitors along the ventricular and subventricular zones in periventricular heterotopia.Hum Mol Genet. 2009 Feb 1;18(3):497-516. doi: 10.1093/hmg/ddn377. Epub 2008 Nov 7. Hum Mol Genet. 2009. PMID: 18996916 Free PMC article.
-
Ablation of MEKK4 kinase activity causes neurulation and skeletal patterning defects in the mouse embryo.Mol Cell Biol. 2005 Oct;25(20):8948-59. doi: 10.1128/MCB.25.20.8948-8959.2005. Mol Cell Biol. 2005. PMID: 16199873 Free PMC article.
-
Filamin A and Filamin B are co-expressed within neurons during periods of neuronal migration and can physically interact.Hum Mol Genet. 2002 Nov 1;11(23):2845-54. doi: 10.1093/hmg/11.23.2845. Hum Mol Genet. 2002. PMID: 12393796
-
Molecular mechanisms of neuronal migration disorders, quo vadis?Curr Mol Med. 2001 Dec;1(6):677-88. doi: 10.2174/1566524013363195. Curr Mol Med. 2001. PMID: 11899256 Review.
Cited by
-
New Insights into Protein Hydroxylation and Its Important Role in Human Diseases.Biochim Biophys Acta. 2016 Dec;1866(2):208-220. doi: 10.1016/j.bbcan.2016.09.004. Epub 2016 Sep 20. Biochim Biophys Acta. 2016. PMID: 27663420 Free PMC article. Review.
-
GPR56 and the developing cerebral cortex: cells, matrix, and neuronal migration.Mol Neurobiol. 2013 Feb;47(1):186-96. doi: 10.1007/s12035-012-8343-0. Epub 2012 Sep 22. Mol Neurobiol. 2013. PMID: 23001883 Free PMC article. Review.
-
Arborization of dendrites by developing neocortical neurons is dependent on primary cilia and type 3 adenylyl cyclase.J Neurosci. 2013 Feb 6;33(6):2626-38. doi: 10.1523/JNEUROSCI.2906-12.2013. J Neurosci. 2013. PMID: 23392690 Free PMC article.
-
Exocyst is involved in polarized cell migration and cerebral cortical development.Proc Natl Acad Sci U S A. 2009 Jul 7;106(27):11342-7. doi: 10.1073/pnas.0904244106. Epub 2009 Jun 19. Proc Natl Acad Sci U S A. 2009. PMID: 19549839 Free PMC article.
-
Formin 2 Regulates Lysosomal Degradation of Wnt-Associated β-Catenin in Neural Progenitors.Cereb Cortex. 2019 May 1;29(5):1938-1952. doi: 10.1093/cercor/bhy073. Cereb Cortex. 2019. PMID: 29659741 Free PMC article.
References
-
- Bai J, Ramos RL, Ackman JB, Thomas AM, Lee RV, LoTurco JJ. RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat Neurosci. 2003;6:1277–1283. - PubMed
-
- Bielas S, Higginbotham H, Koizumi H, Tanaka T, Gleeson JG. Cortical neuronal migration mutants suggest separate but intersecting pathways. Annu Rev Cell Dev Biol. 2004;20:593–618. - PubMed
-
- Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, Schwartz MA, Ginsberg MH. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol. 2001;3:1060–1068. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

