How complete are current yeast and human protein-interaction networks?
- PMID: 17147767
- PMCID: PMC1794583
- DOI: 10.1186/gb-2006-7-11-120
How complete are current yeast and human protein-interaction networks?
Abstract
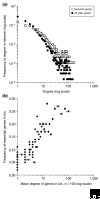
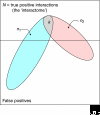
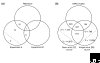
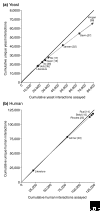
We estimate the full yeast protein-protein interaction network to contain 37,800-75,500 interactions and the human network 154,000-369,000, but owing to a high false-positive rate, current maps are roughly only 50% and 10% complete, respectively. Paradoxically, releasing raw, unfiltered assay data might help separate true from false interactions.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

