Contrasting patterns of sequence divergence and base composition between Drosophila introns and intergenic regions
- PMID: 17148300
- PMCID: PMC1833996
- DOI: 10.1098/rsbl.2006.0521
Contrasting patterns of sequence divergence and base composition between Drosophila introns and intergenic regions
Abstract
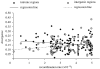
Two non-coding DNA classes, introns and intergenic regions, of Drosophila melanogaster exhibit contrasting evolutionary patterns. GC content is significantly higher in intergenic regions and affects their degree of nucleotide variability. Divergence is positively correlated with recombination rate in intergenic regions, but not in introns. We argue that these differences are due to different selective constraints rather than mutational or recombinational mechanisms.
Figures
References
-
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107 - DOI - PubMed
-
- Chen Y, Stephan W. Weak selection on noncoding gene features. In: Fox C.W, Wolf J.B, editors. Evolutionary genetics—concepts and case studies. Oxford University Press; Oxford, UK: 2006. pp. 133–143.
-
- Galtier N, Bazin E, Bierne N. GC-biased segregation of non-coding polymorphisms in Drosophila. Genetics. 2006;172:221–228. doi:10.1534/genetics.105.046524 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous