PReMod: a database of genome-wide mammalian cis-regulatory module predictions
- PMID: 17148480
- PMCID: PMC1761432
- DOI: 10.1093/nar/gkl879
PReMod: a database of genome-wide mammalian cis-regulatory module predictions
Abstract
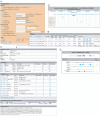
We describe PReMod, a new database of genome-wide cis-regulatory module (CRM) predictions for both the human and the mouse genomes. The prediction algorithm, described previously in Blanchette et al. (2006) Genome Res., 16, 656-668, exploits the fact that many known CRMs are made of clusters of phylogenetically conserved and repeated transcription factors (TF) binding sites. Contrary to other existing databases, PReMod is not restricted to modules located proximal to genes, but in fact mostly contains distal predicted CRMs (pCRMs). Through its web interface, PReMod allows users to (i) identify pCRMs around a gene of interest; (ii) identify pCRMs that have binding sites for a given TF (or a set of TFs) or (iii) download the entire dataset for local analyses. Queries can also be refined by filtering for specific chromosomal regions, for specific regions relative to genes or for the presence of CpG islands. The output includes information about the binding sites predicted within the selected pCRMs, and a graphical display of their distribution within the pCRMs. It also provides a visual depiction of the chromosomal context of the selected pCRMs in terms of neighboring pCRMs and genes, all of which are linked to the UCSC Genome Browser and the NCBI. PReMod: http://genomequebec.mcgill.ca/PReMod.
Figures
References
-
- Aerts S., Van Loo P., Thijs G., Moreau Y., De Moor B. Computational detection of cis-regulatory modules. Bioinformatics. 2003;19:II5–II14. - PubMed
-
- Aerts S., Van Loo P., Moreau Y., De Moor B. A genetic algorithm for the detection of new cis-regulatory modules in sets of coregulated genes. Bioinformatics. 2004;20:1974–1976. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

