Order N algorithm for computation of electrostatic interactions in biomolecular systems
- PMID: 17148613
- PMCID: PMC1748223
- DOI: 10.1073/pnas.0605166103
Order N algorithm for computation of electrostatic interactions in biomolecular systems
Abstract
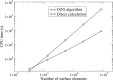
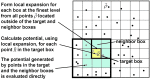
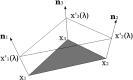
Poisson-Boltzmann electrostatics is a well established model in biophysics; however, its application to large-scale biomolecular processes such as protein-protein encounter is still limited by the efficiency and memory constraints of existing numerical techniques. In this article, we present an efficient and accurate scheme that incorporates recently developed numerical techniques to enhance our computational ability. In particular, a boundary integral equation approach is applied to discretize the linearized Poisson-Boltzmann equation; the resulting integral formulas are well conditioned and are extended to systems with arbitrary numbers of biomolecules. The solution process is accelerated by Krylov subspace methods and a new version of the fast multipole method. In addition to the electrostatic energy, fast calculations of the forces and torques are made possible by using an interpolation procedure. Numerical experiments show that the implemented algorithm is asymptotically optimal O(N) in both CPU time and required memory, and application to the acetylcholinesterase-fasciculin complex is illustrated.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
"New-version-fast-multipole-method" accelerated electrostatic interactions in biomolecular systems.J Comput Phys. 2007 Oct 1;226(2):1348-1366. doi: 10.1016/j.jcp.2007.05.026. J Comput Phys. 2007. PMID: 18379638 Free PMC article.
-
Computation of electrostatic forces between solvated molecules determined by the Poisson-Boltzmann equation using a boundary element method.J Chem Phys. 2005 Jun 1;122(21):214102. doi: 10.1063/1.1924448. J Chem Phys. 2005. PMID: 15974723
-
AFMPB: An Adaptive Fast Multipole Poisson-Boltzmann Solver for Calculating Electrostatics in Biomolecular Systems.Comput Phys Commun. 2010 Jun 1;181(6):1150-1160. doi: 10.1016/j.cpc.2010.02.015. Comput Phys Commun. 2010. PMID: 20532187 Free PMC article.
-
The Poisson-Boltzmann equation for biomolecular electrostatics: a tool for structural biology.J Mol Recognit. 2002 Nov-Dec;15(6):377-92. doi: 10.1002/jmr.577. J Mol Recognit. 2002. PMID: 12501158 Review.
-
Continuum molecular electrostatics, salt effects, and counterion binding--a review of the Poisson-Boltzmann theory and its modifications.Biopolymers. 2008 Feb;89(2):93-113. doi: 10.1002/bip.20877. Biopolymers. 2008. PMID: 17969016 Review.
Cited by
-
Exploring a charge-central strategy in the solution of Poisson's equation for biomolecular applications.Phys Chem Chem Phys. 2013 Jan 7;15(1):129-41. doi: 10.1039/c2cp41894k. Epub 2012 Nov 13. Phys Chem Chem Phys. 2013. PMID: 23147243 Free PMC article.
-
Electrostatic forces in the Poisson-Boltzmann systems.J Chem Phys. 2013 Sep 7;139(9):094106. doi: 10.1063/1.4819471. J Chem Phys. 2013. PMID: 24028101 Free PMC article.
-
An efficient second-order poisson-boltzmann method.J Comput Chem. 2019 May 5;40(12):1257-1269. doi: 10.1002/jcc.25783. Epub 2019 Feb 18. J Comput Chem. 2019. PMID: 30776135 Free PMC article.
-
Accurate solution of multi-region continuum biomolecule electrostatic problems using the linearized Poisson-Boltzmann equation with curved boundary elements.J Comput Chem. 2009 Jan 15;30(1):132-53. doi: 10.1002/jcc.21027. J Comput Chem. 2009. PMID: 18567005 Free PMC article.
-
A new FFT-based algorithm to compute Born radii in the generalized Born theory of biomolecule solvation.J Comput Phys. 2008 Dec 20;227(24):10162-10177. doi: 10.1016/j.jcp.2008.08.015. J Comput Phys. 2008. PMID: 20027201 Free PMC article.
References
-
- Appel AA. SIAM J Sci Stat Comput. 1985;6:85–103.
-
- Barnes J, Hut P. Nature. 1986;324:446–449.
-
- Kuo SS, Altman MD, Bardhan JP, Tidor B, White JK. Proc IEEE/ACM Int Conf Comput Aided Des. 2002:466–473.
-
- Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092.
-
- Greengard L, Rokhlin V. J Comput Phys. 1987;73:325–348.

