Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors
- PMID: 17151109
- PMCID: PMC1865922
- DOI: 10.1128/JVI.02165-06
Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors
Abstract
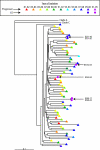
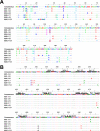
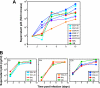
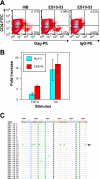
Elite suppressors (ES) are untreated human immunodeficiency virus type 1 (HIV-1)-infected individuals who control viremia to levels below the limit of detection of current assays. The mechanisms involved in this control have not been fully elucidated. Several studies have demonstrated that some ES are infected with defective viruses, but it remains unclear whether others are infected with replication-competent HIV-1. To answer this question, we used a sensitive coculture assay in an attempt to isolate replication-competent virus from a cohort of 10 ES. We successfully cultured six replication-competent isolates from 4 of the 10 ES. The frequency of latently infected cells in these patients was more than a log lower than that seen in patients on highly active antiretroviral therapy with undetectable viral loads. Full-length sequencing of all six isolates revealed no large deletions in any of the genes. A few mutations and small insertions and deletions were found in some isolates, but phenotypic analysis of the affected genes suggested that their function remained intact. Furthermore, all six isolates replicated as well as standard laboratory strains in vitro. The results suggest that some ES are infected with HIV-1 isolates that are fully replication competent and that long-term immunologic control of replication-competent HIV-1 is possible.
Figures






References
-
- Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. - PMC - PubMed
-
- Alexander, L., E. Weiskopf, T. C. Greenough, N. C. Gaddis, M. R. Auerbach, M. H. Malim, S. J. O'Brien, B. D. Walker, J. L. Sullivan, and R. C. Desrosiers. 2000. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J. Virol. 74:4361-4376. - PMC - PubMed
-
- Bailey, J. R., K. G. Lassen, H. C. Yang, T. C. Quinn, S. C. Ray, J. N. Blankson, and R. F. Siliciano. 2006. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J. Virol. 80:4758-4770. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

