Adeno-associated virus site-specific integration is regulated by TRP-185
- PMID: 17151120
- PMCID: PMC1797547
- DOI: 10.1128/JVI.02014-06
Adeno-associated virus site-specific integration is regulated by TRP-185
Abstract
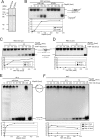
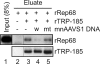
Adeno-associated virus (AAV) integrates site specifically into the AAVS1 locus on human chromosome 19. Although recruitment of the AAV nonstructural protein Rep78/68 to the Rep binding site (RBS) on AAVS1 is thought to be an essential step, the mechanism of the site-specific integration, particularly, how the site of integration is determined, remains largely unknown. Here we describe the identification and characterization of a new cellular regulator of AAV site-specific integration. TAR RNA loop binding protein 185 (TRP-185), previously reported to associate with human immunodeficiency virus type 1 TAR RNA, binds to AAVS1 DNA. Our data suggest that TRP-185 suppresses AAV integration at the AAVS1 RBS and enhances AAV integration into a region downstream of the RBS. TRP-185 bound to Rep68 directly, changing the Rep68 DNA binding property and stimulating Rep68 helicase activity. We present a model in which TRP-185 changes the specificity of the AAV integration site from the RBS to a downstream region by acting as a molecular chaperone that promotes Rep68 complex formation competent for 3'-->5' DNA helicase activity.
Figures





Similar articles
-
A 16bp Rep binding element is sufficient for mediating Rep-dependent integration into AAVS1.J Mol Biol. 2006 Apr 21;358(1):38-45. doi: 10.1016/j.jmb.2006.01.029. Epub 2006 Jan 30. J Mol Biol. 2006. PMID: 16516232
-
A Rep recognition sequence is necessary but not sufficient for nicking of DNA by adeno-associated virus type-2 Rep proteins.Arch Biochem Biophys. 2001 May 15;389(2):271-7. doi: 10.1006/abbi.2001.2348. Arch Biochem Biophys. 2001. PMID: 11339817
-
The transient expression of mRNA coding for Rep protein from AAV facilitates targeted plasmid integration.J Gene Med. 2008 Jan;10(1):42-50. doi: 10.1002/jgm.1118. J Gene Med. 2008. PMID: 18001001
-
Integration of adeno-associated virus (AAV) and recombinant AAV vectors.Annu Rev Genet. 2004;38:819-45. doi: 10.1146/annurev.genet.37.110801.143717. Annu Rev Genet. 2004. PMID: 15568995 Review.
-
Site-specific integration by adeno-associated virus.Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11288-94. doi: 10.1073/pnas.93.21.11288. Proc Natl Acad Sci U S A. 1996. PMID: 8876128 Free PMC article. Review.
Cited by
-
Identification of cellular proteins that interact with the adeno-associated virus rep protein.J Virol. 2009 Jan;83(1):454-69. doi: 10.1128/JVI.01939-08. Epub 2008 Oct 29. J Virol. 2009. PMID: 18971280 Free PMC article.
-
Gene therapy using adeno-associated virus vectors.Clin Microbiol Rev. 2008 Oct;21(4):583-93. doi: 10.1128/CMR.00008-08. Clin Microbiol Rev. 2008. PMID: 18854481 Free PMC article. Review.
References
-
- Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. - PubMed
-
- Costello, E., P. Saudan, E. Winocour, L. Pizer, and P. Beard. 1997. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 16:5943-5954. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

