Crystal structure of apo-calmodulin bound to the first two IQ motifs of myosin V reveals essential recognition features
- PMID: 17151196
- PMCID: PMC1687203
- DOI: 10.1073/pnas.0609436103
Crystal structure of apo-calmodulin bound to the first two IQ motifs of myosin V reveals essential recognition features
Abstract
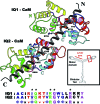
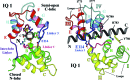
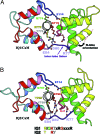
A 2.5-A resolution structure of calcium-free calmodulin (CaM) bound to the first two IQ motifs of the murine myosin V heavy chain reveals an unusual CaM conformation. The C-terminal lobe of each CaM adopts a semi-open conformation that grips the first part of the IQ motif (IQxxxR), whereas the N-terminal lobe adopts a closed conformation that interacts more weakly with the second part of the motif (GxxxR). Variable residues in the IQ motif play a critical role in determining the precise structure of the bound CaM, such that even the consensus residues of different motifs show unique interactions with CaM. This complex serves as a model for the lever arm region of many classes of unconventional myosins, as well as other IQ motif-containing proteins such as neuromodulin and IQGAPs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Trybus KM. Nat Cell Biol. 2005;7:854–856. - PubMed
-
- Sellers JR, Veigel C. Curr Opin Cell Biol. 2006;18:68–73. - PubMed
-
- Wang F, Chen L, Arcucci O, Harvey EV, Bowers B, Xu Y, Hammer JA, III, Sellers JR. J Biol Chem. 2000;275:4329–4335. - PubMed
-
- Wang F, Thirumurugan K, Stafford WF, Hammer JA, III, Knight PJ, Sellers JR. J Biol Chem. 2004;279:2333–2336. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

