Zfp423 controls proliferation and differentiation of neural precursors in cerebellar vermis formation
- PMID: 17151198
- PMCID: PMC1748242
- DOI: 10.1073/pnas.0609184103
Zfp423 controls proliferation and differentiation of neural precursors in cerebellar vermis formation
Abstract
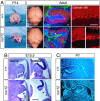
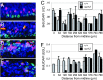
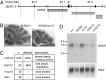
Neural stem cells and progenitors in the developing brain must choose between proliferation with renewal and differentiation. Defects in navigating this choice can result in malformations or cancers, but the genetic mechanisms that shape this choice are not fully understood. We show by positional cloning that the 30-zinc finger transcription factor Zfp423 (OAZ) is required for patterning the development of neuronal and glial precursors in the developing brain, particularly in midline structures. Mutation of Zfp423 results in loss of the corpus callosum, reduction of hippocampus, and a malformation of the cerebellum reminiscent of human Dandy-Walker patients. Within the cerebellum, Zfp423 is expressed in both ventricular and external germinal zones. Loss of Zfp423 results in diminished proliferation by granule cell precursors in the external germinal layer, especially near the midline, and abnormal differentiation and migration of ventricular zone-derived neurons and Bergmann glia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- ten Donkelaar HJ, Lammens M, Wesseling P, Thijssen HO, Renier WO. J Neurol. 2003;250:1025–1036. - PubMed
-
- Grinberg I, Northrup H, Ardinger H, Prasad C, Dobyns WB, Millen KJ. Nat Genet. 2004;36:1053–1055. - PubMed
-
- Kile BT, Hentges KE, Clark AT, Nakamura H, Salinger AP, Liu B, Box N, Stockton DW, Johnson RL, Behringer RR, et al. Nature. 2003;425:81–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

