Onset of the DNA replication checkpoint in the early Drosophila embryo
- PMID: 17151243
- PMCID: PMC1800604
- DOI: 10.1534/genetics.106.065219
Onset of the DNA replication checkpoint in the early Drosophila embryo
Abstract
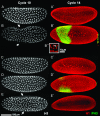
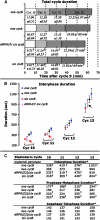
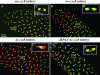
The Drosophila embryo is a promising model for isolating gene products that coordinate S phase and mitosis. We have reported before that increasing maternal Cyclin B dosage to up to six copies (six cycB) increases Cdk1-Cyclin B (CycB) levels and activity in the embryo, delays nuclear migration at cycle 10, and produces abnormal nuclei at cycle 14. Here we show that the level of CycB in the embryo inversely correlates with the ability to lengthen interphase as the embryo transits from preblastoderm to blastoderm stages and defines the onset of a checkpoint that regulates mitosis when DNA replication is blocked with aphidicolin. A screen for modifiers of the six cycB phenotypes identified 10 new suppressor deficiencies. In addition, heterozygote dRPA2 (a DNA replication gene) mutants suppressed only the abnormal nuclear phenotype at cycle 14. Reduction of dRPA2 also restored interphase duration and checkpoint efficacy to control levels. We propose that lowered dRPA2 levels activate Grp/Chk1 to counteract excess Cdk1-CycB activity and restore interphase duration and the ability to block mitosis in response to aphidicolin. Our results suggest an antagonistic interaction between DNA replication checkpoint activation and Cdk1-CycB activity during the transition from preblastoderm to blastoderm cycles.
Figures



References
-
- Blankenship, J. T., and E. Wieschaus, 2001. Two new roles for the Drosophila AP patterning system in early morphogenesis. Development 128: 5129–5138. - PubMed
-
- Blumenthal, A. B., H. J. Kriegstein and D. S. Hogness, 1974. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harbor Symp. Quant. Biol. 38: 205–223. - PubMed
-
- Clarkson, M., and R. Saint, 1999. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 18: 457–462. - PubMed
-
- Costanzo, V., D. Shechter, P. J. Lupardus, K. A. Cimprich, M. Gottesman et al., 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell 11: 203–213. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

