Full-length sequence analysis of SIVmus in wild populations of mustached monkeys (Cercopithecus cephus) from Cameroon provides evidence for two co-circulating SIVmus lineages
- PMID: 17156809
- PMCID: PMC1900428
- DOI: 10.1016/j.virol.2006.10.048
Full-length sequence analysis of SIVmus in wild populations of mustached monkeys (Cercopithecus cephus) from Cameroon provides evidence for two co-circulating SIVmus lineages
Abstract
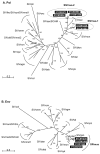
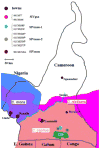
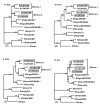
Mustached monkeys (Cercopithecus cephus), which form a significant component of primate bushmeat in west central Africa, are infected with simian immunodeficiency virus (SIVmus). We identified and genetically characterized five new SIVmus strains infecting wild living mustached monkeys from Cameroon. Phylogenetic analysis of partial pol sequences revealed that SIVmus strains form two distinct groups within the clade comprised of lentiviruses isolated from Cercopithecus nictitans (SIVgsn), Cercopithecus mona (SIVmon) and C. cephus (SIVmus). Characterisation of three full-length SIVmus genomes confirmed the presence of two distinct lineages infecting mustached monkeys. These two variants of SIVmus, here designated SIVmus-1 and SIVmus-2, were isolated from animals sharing habitats within the same geographic region. Phylogenetic analyses showed that the diversification of SIVmus, SIVgsn and SIVmon involved inter-lineage recombination, and suggested that one of the SIVmus lineages likely resulted from cross-species transmission and recombination involving SIVmus and an as yet uncharacterized SIV. These results indicate that cross-species transmission and recombination play a major role in the evolution of primate lentiviruses among sympatric primate species.
Figures

References
-
- Aghokeng AF, Liu W, Bibollet-Ruche F, Loul S, Mpoudi-Ngole E, Laurent C, Mwenda JM, Langat DK, Chege GK, McClure HM, Delaporte E, Shaw GM, Hahn BH, Peeters M. Widely varying SIV prevalence rates in naturally infected primate species from Cameroon. Virology. 2006;345:174–189. - PubMed
-
- Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. - PubMed
-
- Bibollet-Ruche F, Bailes E, Gao F, Pourrut X, Barlow KL, Clewley JP, Mwenda JM, Langat DK, Chege GK, McClure HM, Mpoudi-Ngole E, Delaporte E, Peeters M, Shaw GM, Sharp PM, Hahn BH. New simian immunodeficiency virus infecting De Brazza’s monkeys (Cercopithecus neglectus): evidence for a Cercopithecus monkey virus clade. J Virol. 2004;78:7748–7762. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

