Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1
- PMID: 17157258
- PMCID: PMC2859291
- DOI: 10.1016/j.molcel.2006.10.027
Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1
Abstract
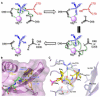
Phosphorylation and dephosphorylation of the C-terminal domain (CTD) of RNA polymerase II (Pol II) represent a critical regulatory checkpoint for transcription. Transcription initiation requires Fcp1/Scp1-mediated dephosphorylation of phospho-CTD. Fcp1 and Scp1 belong to a family of Mg2+ -dependent phosphoserine (P.Ser)/phosphothreonine (P.Thr)-specific phosphatases. We recently showed that Scp1 is an evolutionarily conserved regulator of neuronal gene silencing. Here, we present the X-ray crystal structures of a dominant-negative form of human Scp1 (D96N mutant) bound to mono- and diphosphorylated peptides encompassing the CTD heptad repeat (Y1S2P3T4S5P6S7). Moreover, kinetic and thermodynamic analyses of Scp1-phospho-CTD peptide complexes support the structures determined. This combined structure-function analysis discloses the residues in Scp1 involved in CTD binding and its preferential dephosphorylation of P.Ser5 of the CTD heptad repeat. Moreover, these results provide a template for the design of specific inhibitors of Scp1 for the study of neuronal stem cell development.
Figures


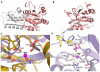
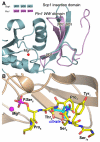
References
-
- Allen KN, Dunaway-Mariano D. Phosphoryl group transfer: evolution of a catalytic scaffold. Trends Biochem. Sci. 2004;29:495–503. - PubMed
-
- Brandts JF, Lin LN. Proline isomerization studied with proteolytic enzymes. Methods Enzymol. 1986;131:107–126. - PubMed
-
- Bregman DB, Pestell RG, Kidd VJ. Cell cycle regulation and RNA polymerase II. Front. Biosci. 2000;5:D244–D257. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

