Different levels of alternative splicing among eukaryotes
- PMID: 17158149
- PMCID: PMC1802581
- DOI: 10.1093/nar/gkl924
Different levels of alternative splicing among eukaryotes
Abstract
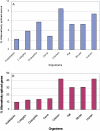
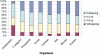
Alternative splicing increases transcriptome and proteome diversification. Previous analyses aiming at comparing the rate of alternative splicing between different organisms provided contradicting results. These contradicting results were attributed to the fact that both analyses were dependent on the expressed sequence tag (EST) coverage, which varies greatly between the tested organisms. In this study we compare the level of alternative splicing among eight different organisms. By employing an EST independent approach we reveal that the percentage of genes and exons undergoing alternative splicing is higher in vertebrates compared with invertebrates. We also find that alternative exons of the skipping type are flanked by longer introns compared to constitutive ones, whereas alternative 5' and 3' splice sites events are generally not. In addition, although the regulation of alternative splicing and sizes of introns and exons have changed during metazoan evolution, intron retention remained the rarest type of alternative splicing, whereas exon skipping is more prevalent and exhibits a slight increase, from invertebrates to vertebrates. The difference in the level of alternative splicing suggests that alternative splicing may contribute greatly to the mammal higher level of phenotypic complexity, and that accumulation of introns confers an evolutionary advantage as it allows increasing the number of alternative splicing forms.
Figures


References
-
- Consortium C.e.S. Genome sequence of the nematode C.elegans: a platform for investigating biology. Science. 1998;282:2012–2018. - PubMed
-
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. - PubMed
-
- Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

