LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro
- PMID: 17158150
- PMCID: PMC1802576
- DOI: 10.1093/nar/gkl885
LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro
Abstract
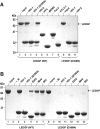
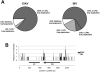
Transcriptional co-activator LEDGF/p75 is the major cellular interactor of HIV-1 integrase (IN), critical to efficient viral replication. In this work, a series of INs from the Betaretrovirus, Gammaretrovirus, Deltaretrovirus, Spumavirus and Lentivirus retroviral genera were tested for interaction with the host factor. None of the non-lentiviral INs possessed detectable affinity for LEDGF in either pull-down or yeast two-hybrid assays. In contrast, all lentiviral INs examined, including those from bovine immunodeficiency virus (BIV), maedi-visna virus (MVV) and equine infectious anemia virus (EIAV) readily interacted with LEDGF. Mutation of Asp-366 to Asn in LEDGF ablated the interaction, suggesting a common mechanism of the host factor recognition by the INs. LEDGF potently stimulated strand transfer activity of divergent lentiviral INs in vitro. Unprecedentedly, in the presence of the host factor, EIAV IN almost exclusively catalyzed concerted integration, whereas HIV-1 IN promoted predominantly half-site integration, and BIV IN was equally active in both types of strand transfer. Concerted BIV and EIAV integration resulted in 5 bp duplications of the target DNA sequences. These results confirm that the interaction with LEDGF is conserved within and limited to Lentivirus and strongly argue that the host factor is intimately involved in the catalysis of lentiviral DNA integration.
Figures



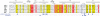
References
-
- Buchen-Osmond C. The universal virus database ICTVDB. Comput. Sci. Eng. 2003;5:16–25.
-
- Grinsztejn B., Nguyen B.Y., Katlama C., Gatell J., Lazzarin A., Vittecoq D., Gonzalez C., Chen J., Isaacs R. The_Protocol_005_Study_Team. The 13th Conference on Retroviruses and Opportunistic Infections; February 5–8, 2006; Denver, CO. 2006.
-
- Hazuda D.J., Felock P., Witmer M., Wolfe A., Stillmock K., Grobler J.A., Espeseth A., Gabryelski L., Schleif W., Blau C., et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. - PubMed
-
- Craigie R. Retroviral DNA integration. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington DC: ASM Press; 2002. pp. 613–630.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

