An efficient method for multi-locus molecular haplotyping
- PMID: 17158153
- PMCID: PMC1802573
- DOI: 10.1093/nar/gkl742
An efficient method for multi-locus molecular haplotyping
Erratum in
- Nucleic Acids Res. 2007;35(9):3165
Abstract
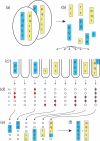
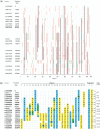
Many methods exist for genotyping--revealing which alleles an individual carries at different genetic loci. A harder problem is haplotyping--determining which alleles lie on each of the two homologous chromosomes in a diploid individual. Conventional approaches to haplotyping require the use of several generations to reconstruct haplotypes within a pedigree, or use statistical methods to estimate the prevalence of different haplotypes in a population. Several molecular haplotyping methods have been proposed, but have been limited to small numbers of loci, usually over short distances. Here we demonstrate a method which allows rapid molecular haplotyping of many loci over long distances. The method requires no more genotypings than pedigree methods, but requires no family material. It relies on a procedure to identify and genotype single DNA molecules, and reconstruction of long haplotypes by a 'tiling' approach. We demonstrate this by resolving haplotypes in two regions of the human genome, harbouring 20 and 105 single-nucleotide polymorphisms, respectively. The method can be extended to reconstruct haplotypes of arbitrary complexity and length, and can make use of a variety of genotyping platforms. We also argue that this method is applicable in situations which are intractable to conventional approaches.
Figures

References
-
- Drysdale C.M., McGraw D.W., Stack C.B., Stephens J.C., Judson R.S., Nandabalan K., Arnold K., Ruano G., Liggett S.B. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl Acad. Sci. USA. 2000;97:10483–10488. - PMC - PubMed
-
- Morris R.W., Kaplan N.L. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet. Epidemiol. 2002;23:221–233. - PubMed
-
- Winkelmann B.R., Hoffmann M.M., Nauck M., Kumar A.M., Nandabalan K., Judson R.S., Boehm B.O., Tall A.R., Ruano G., Marz W. Haplotypes of the cholesteryl ester transfer protein gene predict lipid-modifying response to statin therapy. Pharmacogenomics J. 2003;3:284–296. - PubMed
-
- Clark A.G. The role of haplotypes in candidate gene studies. Genet. Epidemiol. 2004;27:321–333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

