Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos
- PMID: 17158157
- PMCID: PMC1802562
- DOI: 10.1093/nar/gkl1032
Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos
Abstract
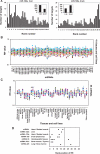
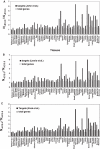
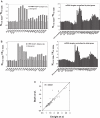
MicroRNAs (miRNAs) are non-coding small RNAs of approximately 22 nt that regulate the gene expression by base pairing with target mRNAs, leading to mRNA cleavage or translational repression. It is currently estimated that miRNAs account for approximately 1% of predicted genes in higher eukaryotic genomes and that up to 30% of genes might be regulated by miRNAs. However, only very few miRNAs have been functionally characterized and the general functions of miRNAs are not globally studied. In this study, we systematically analyzed the expression patterns of miRNA targets using several public microarray profiles. We found that the expression levels of miRNA targets are lower in all mouse and Drosophila tissues than in the embryos. We also found miRNAs more preferentially target ubiquitously expressed genes than tissue-specifically expressed genes. These results support the current suggestion that miRNAs are likely to be largely involved in embryo development and maintaining of tissue identity.
Figures




References
-
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. - PubMed
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Cullen B.R. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. - PubMed
-
- Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell. Biol. 2005;6:376–385. - PubMed
-
- Carmell M.A., Hannon G.J. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004;11:214–218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

