Use of modified U1 snRNAs to inhibit HIV-1 replication
- PMID: 17158512
- PMCID: PMC1802557
- DOI: 10.1093/nar/gkl1022
Use of modified U1 snRNAs to inhibit HIV-1 replication
Abstract
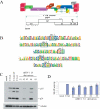
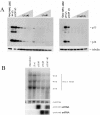
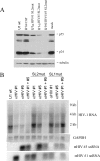
Control of RNA processing plays a central role in regulating the replication of HIV-1, in particular the 3' polyadenylation of viral RNA. Based on the demonstration that polyadenylation of mRNAs can be disrupted by the targeted binding of modified U1 snRNA, we examined whether binding of U1 snRNAs to conserved 10 nt regions within the terminal exon of HIV-1 was able to inhibit viral structural protein expression. In this report, we demonstrate that U1 snRNAs complementary to 5 of the 15 regions targeted result in significant suppression of HIV-1 protein expression and viral replication coincident with loss of viral RNA. Suppression of viral gene expression is dependent upon appropriate assembly of a U1 snRNP particle as mutations of U1 snRNA that affect binding of U1 70K or Sm proteins significantly reduced efficacy. However, constructs lacking U1A binding sites retained significant anti-viral activity. This finding suggests a role for these mutants in situations where the wild-type constructs cause toxic effects. The conserved nature of the sequences targeted and the high efficacy of the constructs suggests that this strategy has significant potential as an HIV therapeutic.
Figures

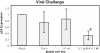
References
-
- O'Reilly M.M., McNally M.T., Beemon K.L. Two strong 5′ (splice sites and competing, suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology. 1995;213:373–385. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

