Molecular dynamics simulation suggests possible interaction patterns at early steps of beta2-microglobulin aggregation
- PMID: 17158575
- PMCID: PMC1796822
- DOI: 10.1529/biophysj.106.098483
Molecular dynamics simulation suggests possible interaction patterns at early steps of beta2-microglobulin aggregation
Abstract
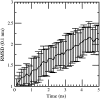
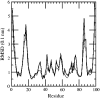
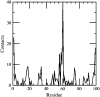
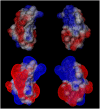
Early events in aggregation of proteins are not easily accessible by experiments. In this work, we perform a 5-ns molecular dynamics simulation of an ensemble of 27 copies of beta(2)-microglobulin in explicit solvent. During the simulation, the formation of intermolecular contacts is observed. The simulation highlights the importance of apical residues and, in particular, of those at the N-terminus end of the molecule. The most frequently found pattern of interaction involves a head-to-head contact arrangement of molecules. Hydrophobic contacts appear to be important for the establishment of long-lived (on the simulation timescale) contacts. Although early events on the pathway to aggregation and fibril formation are not directly related to the end-state of the process, which is reached on a much longer timescale, simulation results are consistent with experimental data and in general with a parallel arrangement of intermolecular beta-strand pairs.
Figures



Similar articles
-
Molecular dynamics simulations and free energy analyses on the dimer formation of an amyloidogenic heptapeptide from human beta2-microglobulin: implication for the protofibril structure.J Mol Biol. 2006 Mar 3;356(4):1049-63. doi: 10.1016/j.jmb.2005.11.087. Epub 2005 Dec 15. J Mol Biol. 2006. PMID: 16403526
-
Molecular dynamics simulation of β₂-microglobulin in denaturing and stabilizing conditions.Proteins. 2011 Mar;79(3):986-1001. doi: 10.1002/prot.22940. Epub 2010 Dec 22. Proteins. 2011. PMID: 21287627
-
Molecular basis for the Cu2+ binding-induced destabilization of beta2-microglobulin revealed by molecular dynamics simulation.Biophys J. 2006 Jun 1;90(11):3865-79. doi: 10.1529/biophysj.105.064444. Epub 2006 Mar 2. Biophys J. 2006. PMID: 16513784 Free PMC article.
-
Coarse-grained models for protein aggregation.Curr Opin Struct Biol. 2011 Apr;21(2):209-20. doi: 10.1016/j.sbi.2011.02.002. Epub 2011 Mar 1. Curr Opin Struct Biol. 2011. PMID: 21371882 Review.
-
Molecular connectivity: intermolecular accessibility and encounter simulation.J Mol Graph Model. 2001;20(1):76-83. doi: 10.1016/s1093-3263(01)00102-4. J Mol Graph Model. 2001. PMID: 11760005 Review.
Cited by
-
Insights from molecular dynamics simulations for computational protein design.Mol Syst Des Eng. 2017 Feb 1;2(1):9-33. doi: 10.1039/C6ME00083E. Epub 2017 Jan 9. Mol Syst Des Eng. 2017. PMID: 28239489 Free PMC article.
-
Understanding the complex mechanisms of β2-microglobulin amyloid assembly.FEBS J. 2011 Oct;278(20):3868-83. doi: 10.1111/j.1742-4658.2011.08186.x. Epub 2011 Jun 13. FEBS J. 2011. PMID: 21595827 Free PMC article. Review.
-
C. elegans expressing human β2-microglobulin: a novel model for studying the relationship between the molecular assembly and the toxic phenotype.PLoS One. 2012;7(12):e52314. doi: 10.1371/journal.pone.0052314. Epub 2012 Dec 21. PLoS One. 2012. PMID: 23284985 Free PMC article.
-
GPU/CPU Algorithm for Generalized Born/Solvent-Accessible Surface Area Implicit Solvent Calculations.J Chem Theory Comput. 2012 Jul 10;8(7):2521-2530. doi: 10.1021/ct3003089. Epub 2012 Jun 15. J Chem Theory Comput. 2012. PMID: 23049488 Free PMC article.
-
A simulated intermediate state for folding and aggregation provides insights into ΔN6 β2-microglobulin amyloidogenic behavior.PLoS Comput Biol. 2014 May 8;10(5):e1003606. doi: 10.1371/journal.pcbi.1003606. eCollection 2014 May. PLoS Comput Biol. 2014. PMID: 24809460 Free PMC article.
References
-
- Yamamoto, S., and F. Gejyo. 2005. Historical background and clinical treatment of dialysis-related amyloidosis. Biochim. Biophys. Acta. 1753:4–10. - PubMed
-
- Radford, S. E., W. S. Gosal, and G. W. Platt. 2005. Towards an understanding of the structural molecular mechanism of β2-microglobulin amyloid formation in vitro. Biochim. Biophys. Acta. 1753:51–63. - PubMed
-
- Esposito, G., A. Corazza, P. Viglino, G. Verdone, F. Pettirossi, F. Fogolari, A. Makek, S. Giorgetti, P. Mangione, M. Stoppini, and V. Bellotti. 2005. Solution structure of β2-microglobulin and insights into fibrillogenesis. Biochim. Biophys. Acta. 1753:76–84. - PubMed
-
- Kardos, J., D. Okuno, T. Kawai, Y. Hagihara, N. Yumoto, T. Kitagawa, P. Zavodszky, H. Naiki, and Y. Goto. 2005. Structural studies reveal that the diverse morphology of β2-microglobulin aggregates is a reflection of different molecular architectures. Biochim. Biophys. Acta. 1753:108–120. - PubMed
-
- Corazza, A., F. Pettirossi, P. Viglino, G. Verdone, J. Garcia, P. Dumy, S. Giorgetti, P. Mangione, S. Raimondi, M. Stoppini, V. Bellotti, and G. Esposito. 2004. Properties of some variants of human β2-microglobulin and amyloidogenesis. J. Biol. Chem. 279:9176–9189. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

