Whole-genome analysis of the methyl tert-butyl ether-degrading beta-proteobacterium Methylibium petroleiphilum PM1
- PMID: 17158667
- PMCID: PMC1855728
- DOI: 10.1128/JB.01259-06
Whole-genome analysis of the methyl tert-butyl ether-degrading beta-proteobacterium Methylibium petroleiphilum PM1
Erratum in
- J Bacteriol. 2007 Jul;189(13):4973
Abstract
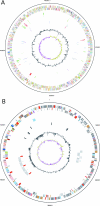
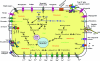
Methylibium petroleiphilum PM1 is a methylotroph distinguished by its ability to completely metabolize the fuel oxygenate methyl tert-butyl ether (MTBE). Strain PM1 also degrades aromatic (benzene, toluene, and xylene) and straight-chain (C(5) to C(12)) hydrocarbons present in petroleum products. Whole-genome analysis of PM1 revealed an approximately 4-Mb circular chromosome and an approximately 600-kb megaplasmid, containing 3,831 and 646 genes, respectively. Aromatic hydrocarbon and alkane degradation, metal resistance, and methylotrophy are encoded on the chromosome. The megaplasmid contains an unusual t-RNA island, numerous insertion sequences, and large repeated elements, including a 40-kb region also present on the chromosome and a 29-kb tandem repeat encoding phosphonate transport and cobalamin biosynthesis. The megaplasmid also codes for alkane degradation and was shown to play an essential role in MTBE degradation through plasmid-curing experiments. Discrepancies between the insertion sequence element distribution patterns, the distributions of best BLASTP hits among major phylogenetic groups, and the G+C contents of the chromosome (69.2%) and plasmid (66%), together with comparative genome hybridization experiments, suggest that the plasmid was recently acquired and apparently carries the genetic information responsible for PM1's ability to degrade MTBE. Comparative genomic hybridization analysis with two PM1-like MTBE-degrading environmental isolates (approximately 99% identical 16S rRNA gene sequences) showed that the plasmid was highly conserved (ca. 99% identical), whereas the chromosomes were too diverse to conduct resequencing analysis. PM1's genome sequence provides a foundation for investigating MTBE biodegradation and exploring the genetic regulation of multiple biodegradation pathways in M. petroleiphilum and other MTBE-degrading beta-proteobacteria.
Figures


References
-
- Afolabi, P. R., F. Mohammed, K. Amaratunga, O. Majekodunmi, S. L. Dales, R. Gill, D. Thompson, J. B. Cooper, S. P. Wood, P. M. Goodwin, and C. Anthony. 2001. Site-directed mutagenesis and X-ray crystallography of the PQQ-containing quinoprotein methanol dehydrogenase and its electron acceptor, cytochrome cL. Biochemistry 40:9799-9809. - PubMed
-
- Asperger, O., A. Naumann, and H.-P. Kleber. 1984. Inducibility of cytochrome P-450 in Acinetobacter calcoaceticus by n-alkanes. Appl. Microbiol. Biotechnol. 19:3948-4403.
-
- Atkinson, R. 1995. Gas phase tropospheric chemistry of organic compounds. Environ. Sci. Technol. 4:65-89.
-
- Atomi, H. 2002. Microbial enzymes involved in carbon dioxide fixation. J. Biosci. Bioeng. 94:497-505. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Stuhl. 1987. Current protocols in molecular biology. Wiley, New York, NY.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

