The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity
- PMID: 17158748
- PMCID: PMC1686608
- DOI: 10.1101/gad.396106
The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity
Abstract
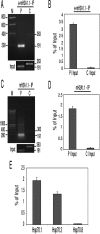
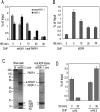
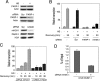
The histone variant mH2A is believed to be involved in transcriptional repression, but how it exerts its function remains elusive. By using chromatin immunoprecipitation and tandem affinity immunopurification of the mH2A1.1 nucleosome complex, we identified numerous genes with promoters containing mH2A1.1 nucleosomes. In particular, the promoters of the inducible Hsp70.1 and Hsp70.2 genes, but not that of the constitutively expressed Hsp70.8, were highly enriched in mH2A1.1. PARP-1 was identified as a part of the mH2A1.1 nucleosome complex and was found to be associated with the Hsp70.1 promoter. A specific interaction between mH2A1.1 and PARP-1 was demonstrated and found to be associated with inactivation of PARP-1 enzymatic activity. Heat shock released both mH2A1.1 and PARP-1 from the Hsp70.1 promoter and activated PARP-1 automodification activity. The data we present point to a novel mechanism for control of Hsp70.1 gene transcription. mH2A1.1 recruits PARP-1 to the promoter, thereby inactivating it. Upon heat shock, the Hsp70.1 promoter-bound PARP-1 is released to activate transcription through ADP-ribosylation of other Hsp70.1 promoter-bound proteins.
Figures




References
-
- Abbott, D.W., Chadwick, B.P., Thambirajah, A.A., Ausio, J. Beyond the Xi: MacroH2A chromatin distribution and post-translational modification in an avian system. J. Biol. Chem. 2005;280:16437–16445. - PubMed
-
- Allen, M.D., Buckle, A.M., Cordell, S.C., Lowe, J., Bycroft, M. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 2003;330:503–511. - PubMed
-
- Ame, J.C., Spenlehauer, C., de Murcia, G. The PARP superfamily. Bioessays. 2004;26:882–893. - PubMed
-
- Angelov, D., Molla, A., Perche, P.Y., Hans, F., Cote, J., Khochbin, S., Bouvet, P., Dimitrov, S. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell. 2003;11:1033–1041. - PubMed
-
- Chadwick, B.P., Willard, H.F. Histone H2A variants and the inactive X chromosome: Identification of a second macroH2A variant. Hum. Mol. Genet. 2001;10:1101–1113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
