Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm
- PMID: 17158916
- PMCID: PMC2997529
- DOI: 10.1242/jcs.03292
Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm
Abstract
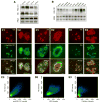
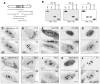
Nucleostemin plays an essential role in maintaining the continuous proliferation of stem cells and cancer cells. The movement of nucleostemin between the nucleolus and the nucleoplasm provides a dynamic way to partition the nucleostemin protein between these two compartments. Here, we show that nucleostemin contains two nucleolus-targeting regions, the basic and the GTP-binding domains, that exhibit a short and a long nucleolar retention time, respectively. In a GTP-unbound state, the nucleolus-targeting activity of nucleostemin is blocked by a mechanism that traps its intermediate domain in the nucleoplasm. A nucleostemin-interacting protein, RSL1D1, was identified that contains a ribosomal L1-domain. RSL1D1 co-resides with nucleostemin in the same subnucleolar compartment, unlike the B23 and fibrillarin, and displays a longer nucleolar residence time than nucleostemin. It interacts with both the basic and the GTP-binding domains of nucleostemin through a non-nucleolus-targeting region. Overexpression of the nucleolus-targeting domain of RSL1D1 alone disperses nucleolar nucleostemin. Loss of RSL1D1 expression reduces the compartmental size and amount of nucleostemin in the nucleolus. Our work reveals that the partitioning of nucleostemin employs complex mechanisms involving both nucleolar and nucleoplasmic components, and provides insight into the post-translational regulation of its activity.
Figures








References
-
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. - PubMed
-
- Baddoo M, Hill K, Wilkinson R, Gaupp D, Hughes C, Kopen GC, Phinney DG. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J Cell Biochem. 2003;89:1235–1249. - PubMed
-
- Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6:665–672. - PubMed
-
- Carmo-Fonseca M, Mendes-Soares L, Campos I. To be or not to be in the nucleolus. Nat Cell Biol. 2000;2:E107–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

