Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts
- PMID: 17158958
- PMCID: PMC2064699
- DOI: 10.1083/jcb.200609172
Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts
Abstract
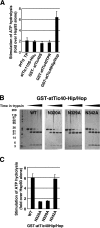
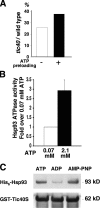
Three components of the chloroplast protein translocon, Tic110, Hsp93 (ClpC), and Tic40, have been shown to be important for protein translocation across the inner envelope membrane into the stroma. We show the molecular interactions among these three components that facilitate processing and translocation of precursor proteins. Transit-peptide binding by Tic110 recruits Tic40 binding to Tic110, which in turn causes the release of transit peptides from Tic110, freeing the transit peptides for processing. The Tic40 C-terminal domain, which is homologous to the C terminus of cochaperones Sti1p/Hop and Hip but with no known function, stimulates adenosine triphosphate hydrolysis by Hsp93. Hsp93 dissociates from Tic40 in the presence of adenosine diphosphate, suggesting that Tic40 functions as an adenosine triphosphatase activation protein for Hsp93. Our data suggest that chloroplasts have evolved the Tic40 cochaperone to increase the efficiency of precursor processing and translocation.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

