Adaptation of a diverse simian immunodeficiency virus population to a new host is revealed through a systematic approach to identify amino acid sites under selection
- PMID: 17159231
- PMCID: PMC7107550
- DOI: 10.1093/molbev/msl194
Adaptation of a diverse simian immunodeficiency virus population to a new host is revealed through a systematic approach to identify amino acid sites under selection
Abstract
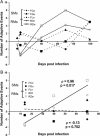
Simian immunodeficiency viruses (SIV) have had considerable success at crossing species barriers; both human immunodeficiency virus (HIV)-1 and HIV-2 have been transmitted on multiple occasions from SIV-infected natural host species. However, the precise evolutionary and ecological mechanisms characterizing a successful cross-species transmission event remain to be elucidated. Here, in addition to expanding and clarifying our previous description of the adaptation of a diverse, naturally occurring SIVsm inoculum to a new rhesus macaque host, we present an analytical framework for understanding the selective forces driving viral adaptation to a new host. A preliminary analysis of large-scale changes in virus population structure revealed that viruses replicating in the macaques were subject to increasing levels of selection through day 70 postinfection (p.i.), whereas contemporaneous viruses in the mangabeys remained similar to the source inoculum. Three different site-by-site methods were employed to identify the amino acid sites responsible for this macaque-specific selection. Of 124 amino acid sites analyzed, 3 codons in V2, a 2-amino acid shift in an N-linked glycosylation site, and variation at 2 sites in the highly charged region were consistently evolving under either directional or diversifying selection at days 40 and 70 p.i. This strong macaque-specific selection on the V2 loop underscores the importance of this region in the adaptation of SIVsm to rhesus macaques. Due to the extreme viral diversity already extant in the naturally occurring viral inoculum, we employed a broad range of phylogenetic and numerical tools in order to distinguish the signatures of past episodes of selection in viral sequences from more recent selection pressures.
Figures


Similar articles
-
Tracking the Emergence of Host-Specific Simian Immunodeficiency Virus env and nef Populations Reveals nef Early Adaptation and Convergent Evolution in Brain of Naturally Progressing Rhesus Macaques.J Virol. 2015 Aug;89(16):8484-96. doi: 10.1128/JVI.01010-15. Epub 2015 Jun 3. J Virol. 2015. PMID: 26041280 Free PMC article.
-
Different evolution of simian immunodeficiency virus in a natural host and a new host.Virology. 1998 Jul 20;247(1):41-50. doi: 10.1006/viro.1998.9217. Virology. 1998. PMID: 9683570
-
Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop.J Virol. 1996 Jun;70(6):3617-27. doi: 10.1128/JVI.70.6.3617-3627.1996. J Virol. 1996. PMID: 8648696 Free PMC article.
-
Phylogeny and natural history of the primate lentiviruses, SIV and HIV.Curr Opin Genet Dev. 1995 Dec;5(6):798-806. doi: 10.1016/0959-437x(95)80014-v. Curr Opin Genet Dev. 1995. PMID: 8745080 Review.
-
Naturally SIV-infected sooty mangabeys: are we closer to understanding why they do not develop AIDS?J Med Primatol. 2005 Oct;34(5-6):243-52. doi: 10.1111/j.1600-0684.2005.00122.x. J Med Primatol. 2005. PMID: 16128919 Review.
Cited by
-
Potent antibody-mediated neutralization and evolution of antigenic escape variants of simian immunodeficiency virus strain SIVmac239 in vivo.J Virol. 2008 Oct;82(19):9739-52. doi: 10.1128/JVI.00871-08. Epub 2008 Jul 30. J Virol. 2008. PMID: 18667507 Free PMC article.
-
APOBEC3G polymorphism as a selective barrier to cross-species transmission and emergence of pathogenic SIV and AIDS in a primate host.PLoS Pathog. 2013;9(10):e1003641. doi: 10.1371/journal.ppat.1003641. Epub 2013 Oct 3. PLoS Pathog. 2013. PMID: 24098115 Free PMC article.
-
Into the wild: simian immunodeficiency virus (SIV) infection in natural hosts.Trends Immunol. 2008 Sep;29(9):419-28. doi: 10.1016/j.it.2008.05.004. Trends Immunol. 2008. PMID: 18676179 Free PMC article. Review.
References
-
- Apetrei C, Lerche NW, Pandrea I, Gormus B, Silvestri G, Kaur A, Robertson DL, Hardcastle J, Lackner AA, Marx PA. Kuru experiments triggered the emergence of pathogenic SIVmac. Aids. 2006;20(3):317–321. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

