Characterization of GATA-1(+) hemangioblastic cells in the mouse embryo
- PMID: 17159898
- PMCID: PMC1782368
- DOI: 10.1038/sj.emboj.7601480
Characterization of GATA-1(+) hemangioblastic cells in the mouse embryo
Abstract
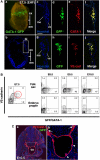
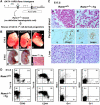
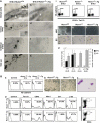
Hemangioblasts are thought to be one of the sources of hematopoietic progenitors, yet little is known about their localization and fate in the mouse embryo. We show here that a subset of cells co-expressing the hematopoietic marker GATA-1 and the endothelial marker VE-cadherin localize on the yolk sac blood islands at embryonic day 7.5. Clonal analysis demonstrated that GATA-1(+) cells isolated from E7.0-7.5 embryos include a common precursor for hematopoietic and endothelial cells. Moreover, this precursor possesses primitive and definitive hematopoietic bipotential. By using a transgenic complementation rescue approach, GATA-1(+) cell-derived progenitors were selectively restored in Runx1-deficient mice. In the rescued mice, definitive erythropoiesis was recovered but the rescued progenitors did not display multilineage hematopoiesis or intra-aortic hematopoietic clusters. These results provide evidence of the presence of GATA-1(+) hemangioblastic cells in the extra-embryonic region and also their functional contribution to hematopoiesis in the embryo.
Figures



References
-
- Adya N, Castilla LH, Liu PP (2000) Function of CBFβ/Bro proteins. Semin Cell Dev Biol 11: 361–368 - PubMed
-
- Cai ZL, de Bruijn M, Ma XQ, Dortland B, Luteijn T, Downing JR, Dzierzak E (2000) Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity 13: 423–431 - PubMed
-
- Choi K (2002) The hemangioblast: a common progenitor of hematopoietic and endothelial cells. J Hematother Stem Cell Res 11: 91–101 - PubMed
-
- Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G (1998) A common precursor for hematopoietic and endothelial cells. Development 125: 725–732 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

