Structural and biochemical characterization of the yeast exosome component Rrp40
- PMID: 17159918
- PMCID: PMC1796750
- DOI: 10.1038/sj.embor.7400856
Structural and biochemical characterization of the yeast exosome component Rrp40
Abstract
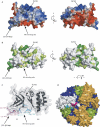
The exosome is a protein complex that is important in both degradation and 3'-processing of eukaryotic RNAs. We present the crystal structure of the Rrp40 exosome subunit from Saccharomyces cerevisiae at a resolution of 2.2 A. The structure comprises an S1 domain and an unusual KH (K homology) domain. Close packing of the S1 and KH domains is stabilized by a GxNG sequence, which is uniquely conserved in exosome KH domains. Nuclear magnetic resonance data reveal the presence of a manganese-binding site at the interface of the two domains. Isothermal titration calorimetry shows that Rrp40 and archaeal Rrp4 alone have very low intrinsic affinity for RNA. The affinity of an archaeal core exosome for RNA is significantly increased in the presence of the S1-KH subunit Rrp4, indicating that multiple subunits might contribute to cooperative binding of RNA substrates by the exosome.
Figures



References
-
- Butler JS (2002) The yin and yang of the exosome. Trends Cell Biol 12: 90–96 - PubMed
-
- Büttner K, Wenig K, Hopfner KP (2005) Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell 20: 461–471 - PubMed
-
- Chekanova JA, Shaw RJ, Wills MA, Belostotsky DA (2000) Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J Biol Chem 275: 33158–33166 - PubMed
-
- Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX Pipes. J Biomol NMR 6: 277–293 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

