A comprehensive analysis of common copy-number variations in the human genome
- PMID: 17160897
- PMCID: PMC1785303
- DOI: 10.1086/510560
A comprehensive analysis of common copy-number variations in the human genome
Abstract
Segmental copy-number variations (CNVs) in the human genome are associated with developmental disorders and susceptibility to diseases. More importantly, CNVs may represent a major genetic component of our phenotypic diversity. In this study, using a whole-genome array comparative genomic hybridization assay, we identified 3,654 autosomal segmental CNVs, 800 of which appeared at a frequency of at least 3%. Of these frequent CNVs, 77% are novel. In the 95 individuals analyzed, the two most diverse genomes differed by at least 9 Mb in size or varied by at least 266 loci in content. Approximately 68% of the 800 polymorphic regions overlap with genes, which may reflect human diversity in senses (smell, hearing, taste, and sight), rhesus phenotype, metabolism, and disease susceptibility. Intriguingly, 14 polymorphic regions harbor 21 of the known human microRNAs, raising the possibility of the contribution of microRNAs to phenotypic diversity in humans. This in-depth survey of CNVs across the human genome provides a valuable baseline for studies involving human genetics.
Figures


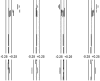
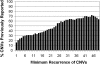




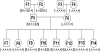
Comment in
-
Copy-number variations and human disease.Am J Hum Genet. 2007 Aug;81(2):414-5; author reply 415. doi: 10.1086/519220. Am J Hum Genet. 2007. PMID: 17668391 Free PMC article. No abstract available.
-
Numbers of copy-number variations and false-negative rates will be underestimated if we do not account for the dependence between repeated experiments.Am J Hum Genet. 2007 Aug;81(2):418-20; author reply 420-1. doi: 10.1086/519393. Am J Hum Genet. 2007. PMID: 17668395 Free PMC article. No abstract available.
-
Estimating prevalence, false-positive rate, and false-negative rate with use of repeated testing when true responses are unknown.Am J Hum Genet. 2007 Nov;81(5):1111-3. doi: 10.1086/521582. Am J Hum Genet. 2007. PMID: 17924351 Free PMC article. No abstract available.
References
Web Resources
-
- BACPAC Resources, http://bacpac.chori.org/genomicRearrays.php (for UCSC May 2004 mapping annotations)
-
- Database of Genomic Variants, http://projects.tcag.ca/variation/
-
- Eisen Lab: Software, http://rana.lbl.gov/EisenSoftware.htm (for Cluster and Treeview)
-
- Gene Expression Omnibus (GEO), http://www.ncbi.nlm.nih.gov/geo/
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

