Structural analysis of the plakin domain of bullous pemphigoid antigen1 (BPAG1) suggests that plakins are members of the spectrin superfamily
- PMID: 17161423
- PMCID: PMC1850962
- DOI: 10.1016/j.jmb.2006.11.036
Structural analysis of the plakin domain of bullous pemphigoid antigen1 (BPAG1) suggests that plakins are members of the spectrin superfamily
Abstract
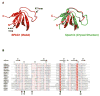
Bullous pemphigoid antigen 1 (BPAG1) is a member of the plakin family of proteins. The plakins are multi-domain proteins that have been shown to interact with microtubules, actin filaments and intermediate filaments, as well as proteins found in cellular junctions. These interactions are mediated through different domains on the plakins. The interactions between plakins and components of specialized cell junctions such as desmosomes and hemidesmosomes are mediated through the so-called plakin domain, which is a common feature of the plakins. We report the crystal structure of a stable fragment from BPAG1, residues 226-448, defined by limited proteolysis of the whole plakin domain. The structure, determined by single-wavelength anomalous diffraction phasing from a selenomethionine-substituted crystal at 3.0 A resolution, reveals a tandem pair of triple helical bundles closely related to spectrin repeats. Based on this structure and analysis of sequence conservation, we propose that the architecture of plakin domains is defined by two pairs of spectrin repeats interrupted by a putative Src-Homology 3 (SH3) domain.
Figures








Similar articles
-
Crystal structure of a rigid four-spectrin-repeat fragment of the human desmoplakin plakin domain.J Mol Biol. 2011 Jun 24;409(5):800-12. doi: 10.1016/j.jmb.2011.04.046. Epub 2011 Apr 22. J Mol Biol. 2011. PMID: 21536047 Free PMC article.
-
Plakins in development and disease.Exp Cell Res. 2007 Jun 10;313(10):2189-203. doi: 10.1016/j.yexcr.2007.03.039. Epub 2007 Apr 19. Exp Cell Res. 2007. PMID: 17499243 Review.
-
The structure of a tandem pair of spectrin repeats of plectin reveals a modular organization of the plakin domain.J Mol Biol. 2007 May 18;368(5):1379-91. doi: 10.1016/j.jmb.2007.02.090. Epub 2007 Mar 7. J Mol Biol. 2007. PMID: 17397861
-
Bpag1 localization to actin filaments and to the nucleus is regulated by its N-terminus.J Cell Sci. 2003 Nov 15;116(Pt 22):4543-55. doi: 10.1242/jcs.00764. J Cell Sci. 2003. PMID: 14576348
-
Plakins in striated muscle.Muscle Nerve. 2010 Mar;41(3):299-308. doi: 10.1002/mus.21472. Muscle Nerve. 2010. PMID: 19768769 Review.
Cited by
-
Plakins, a versatile family of cytolinkers: roles in skin integrity and in human diseases.J Invest Dermatol. 2014 Apr;134(4):885-894. doi: 10.1038/jid.2013.498. Epub 2013 Dec 19. J Invest Dermatol. 2014. PMID: 24352042 Review.
-
The structure of the ankyrin-binding site of beta-spectrin reveals how tandem spectrin-repeats generate unique ligand-binding properties.Blood. 2009 May 28;113(22):5377-84. doi: 10.1182/blood-2008-10-184291. Epub 2009 Jan 23. Blood. 2009. PMID: 19168783 Free PMC article.
-
Advances and perspectives of the architecture of hemidesmosomes: lessons from structural biology.Cell Adh Migr. 2009 Oct-Dec;3(4):361-4. doi: 10.4161/cam.3.4.9525. Epub 2009 Oct 16. Cell Adh Migr. 2009. PMID: 19736524 Free PMC article. Review.
-
Crystal structure of a rigid four-spectrin-repeat fragment of the human desmoplakin plakin domain.J Mol Biol. 2011 Jun 24;409(5):800-12. doi: 10.1016/j.jmb.2011.04.046. Epub 2011 Apr 22. J Mol Biol. 2011. PMID: 21536047 Free PMC article.
-
Mammalian Plakins, Giant Cytolinkers: Versatile Biological Functions and Roles in Cancer.Int J Mol Sci. 2018 Mar 24;19(4):974. doi: 10.3390/ijms19040974. Int J Mol Sci. 2018. PMID: 29587367 Free PMC article. Review.
References
-
- Jefferson JJ, Leung CL, Liem RK. Plakins: goliaths that link cell junctions and the cytoskeleton. Nat Rev Mol Cell Biol. 2004;5:542–53. - PubMed
-
- Leung CL, Green KJ, Liem RK. Plakins: a family of versatile cytolinker proteins. Trends Cell Biol. 2002;12:37–45. - PubMed
-
- Choi HJ, Park-Snyder S, Pascoe LT, Green KJ, Weis WI. Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure. Nat Struct Biol. 2002;9:612–20. - PubMed
-
- Getsios S, Huen AC, Green KJ. Working out the strength and flexibility of desmosomes. Nat Rev Mol Cell Biol. 2004;5:271–81. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

