Structural and functional comparison of native pentameric, denatured monomeric and biotinylated C-reactive protein
- PMID: 17163961
- PMCID: PMC2265887
- DOI: 10.1111/j.1365-2567.2006.02516.x
Structural and functional comparison of native pentameric, denatured monomeric and biotinylated C-reactive protein
Abstract
There are many controversies surrounding the biological activities of native C-reactive protein (nCRP) and its various modified forms such as monomerized and biotinylated CRP (mCRP and bCRP). No simple methods have been described to distinguish among these forms. By adapting established electrophoresis methods, we have developed a useful quality control method with which we have investigated the structural and functional characteristics of these forms of CRP. Under all electrophoresis conditions, biotinylation altered the electrophoretic mobility of CRP. nCRP was sensitive to sodium dodecyl sulphate (SDS)-induced monomerization, and only mCRP was susceptible to digestion by trypsin or neutrophil-derived serine proteases. bCRP and mCRP but not nCRP bound to cells, suggesting that chemical modification by biotin and denaturation had altered the structural integrity of CRP. Neither nCRP nor mCRP had the ability to induce secretion of chemokines, nor did they increase intracellular adhesion molecule 1 (ICAM-1) expression in endothelial cells.
Figures
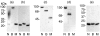
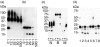

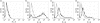
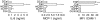
References
-
- Volanakis JE. Human C-reactive protein: expression, structure, and function. Mol Immunol. 2001;38:189–97. - PubMed
-
- Myles DA, Rule SA, DeLucas LJ, et al. Rotation function studies of human C-reactive protein. J Mol Biol. 1990;216:491–6. - PubMed
-
- Shrive AK, Cheetham GM, Holden D, et al. Three dimensional structure of human C-reactive protein. Nat Struct Biol. 1996;3:346–54. - PubMed
-
- Thompson D, Pepys MB, Wood SP. The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure. 1999;7:169–77. - PubMed
-
- Potempa LA, Maldonado BA, Laurent P, Zemel ES, Gewurz H. Antigenic, electrophoretic and binding alterations of human C-reactive protein modified selectively in the absence of calcium. Mol Immunol. 1983;20:1165–75. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

