Comparative transcriptional profiling of the lung reveals shared and distinct features of Streptococcus pneumoniae and influenza A virus infection
- PMID: 17163962
- PMCID: PMC2265881
- DOI: 10.1111/j.1365-2567.2006.02514.x
Comparative transcriptional profiling of the lung reveals shared and distinct features of Streptococcus pneumoniae and influenza A virus infection
Abstract
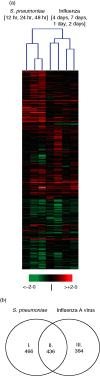
Pneumonia is the most common cause of death from infectious disease in the western hemisphere. Pathophysiological and protective processes are initiated by pattern recognition of microbial structures. To provide the molecular framework for a better understanding of processes relevant to host defence in pneumonia, we performed pulmonary transcriptome analysis in mice infected with the major bacterial and viral agents of community-acquired pneumonia, Streptococcus pneumoniae and influenza A virus. We detected differential expression of 1300 genes after infection with either pathogen. Of these, approximately 36% or 30% were specific for pneumococcal or influenza infection, respectively, yielding pathogen-specific as well as shared inflammatory transcriptional signatures. These results not only reveal a differential response on the cytokine and chemokine levels but also emphasize the important role of genes implicated in regulation and fine tuning of inflammation. As one, albeit unexpected, key feature of pneumococcal pneumonia we discovered down-regulation of B-cell responses, probably reflecting a pneumococcal virulence strategy. The pathophysiological consequences of influenza A virus infection were reflected by the emerging protective T-cell response and differential induction of genes involved in tissue regeneration and proliferation. These data provide new insights into pathogenesis of the most common forms of pneumonia, highlighting the value of transcriptional profiling for the elucidation of underlying mechanisms.
Figures


Similar articles
-
Dysregulated macrophage-inflammatory protein-2 expression drives illness in bacterial superinfection of influenza.J Immunol. 2010 Feb 15;184(4):2001-13. doi: 10.4049/jimmunol.0903304. Epub 2010 Jan 11. J Immunol. 2010. PMID: 20065113
-
Alteration of gene expression in human middle ear epithelial cells induced by influenza A virus and its implication for the pathogenesis of otitis media.Microb Pathog. 2004 Oct;37(4):193-204. doi: 10.1016/j.micpath.2004.06.012. Microb Pathog. 2004. PMID: 15458780
-
Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia.PLoS Pathog. 2006 Jun;2(6):e53. doi: 10.1371/journal.ppat.0020053. Epub 2006 Jun 9. PLoS Pathog. 2006. PMID: 16789835 Free PMC article.
-
Of mice and men: innate immunity in pneumococcal pneumonia.Int J Antimicrob Agents. 2010 Feb;35(2):107-13. doi: 10.1016/j.ijantimicag.2009.10.002. Epub 2009 Dec 14. Int J Antimicrob Agents. 2010. PMID: 20005681 Review.
-
Inflammation as a Modulator of Host Susceptibility to Pulmonary Influenza, Pneumococcal, and Co-Infections.Front Immunol. 2020 Feb 11;11:105. doi: 10.3389/fimmu.2020.00105. eCollection 2020. Front Immunol. 2020. PMID: 32117259 Free PMC article. Review.
Cited by
-
Clinical, immunological and bacteriological characteristics of H7N9 patients nosocomially co-infected by Acinetobacter Baumannii: a case control study.BMC Infect Dis. 2018 Dec 14;18(1):664. doi: 10.1186/s12879-018-3447-4. BMC Infect Dis. 2018. PMID: 30551738 Free PMC article.
-
Systems-based candidate genes for human response to influenza infection.Infect Genet Evol. 2009 Dec;9(6):1148-57. doi: 10.1016/j.meegid.2009.07.006. Epub 2009 Jul 30. Infect Genet Evol. 2009. PMID: 19647099 Free PMC article. Review.
-
Mincle and human B cell function.J Autoimmun. 2012 Dec;39(4):315-22. doi: 10.1016/j.jaut.2012.04.004. Epub 2012 Jun 12. J Autoimmun. 2012. PMID: 22698596 Free PMC article.
-
The innate immune response to Streptococcus pneumoniae in the lung depends on serotype and host response.Vaccine. 2011 Oct 19;29(45):8002-11. doi: 10.1016/j.vaccine.2011.08.064. Epub 2011 Aug 22. Vaccine. 2011. PMID: 21864623 Free PMC article.
-
Differential pulmonary transcriptomic profiles in murine lungs infected with low and highly virulent influenza H3N2 viruses reveal dysregulation of TREM1 signaling, cytokines, and chemokines.Funct Integr Genomics. 2012 Mar;12(1):105-17. doi: 10.1007/s10142-011-0247-y. Epub 2011 Aug 28. Funct Integr Genomics. 2012. PMID: 21874528
References
-
- Shorr AF. Preventing pneumonia: the role for pneumococcal and influenza vaccines. Clin Chest Med. 2005;26:123–34. - PubMed
-
- Oliveira EC, Lee B, Colice GL. Influenza in the intensive care unit. J Intensive Care Med. 2003;18:80–91. - PubMed
-
- Novak R, Tuomanen E. Pathogenesis of pneumococcal pneumonia. Semin Respir Infect. 1999;14:209–17. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

