Influence of sequence identity and unique breakpoints on the frequency of intersubtype HIV-1 recombination
- PMID: 17164002
- PMCID: PMC1764423
- DOI: 10.1186/1742-4690-3-91
Influence of sequence identity and unique breakpoints on the frequency of intersubtype HIV-1 recombination
Abstract
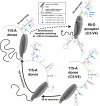
Background: HIV-1 recombination between different subtypes has a major impact on the global epidemic. The generation of these intersubtype recombinants follows a defined set of events starting with dual infection of a host cell, heterodiploid virus production, strand transfers during reverse transcription, and then selection. In this study, recombination frequencies were measured in the C1-C4 regions of the envelope gene in the presence (using a multiple cycle infection system) and absence (in vitro reverse transcription and single cycle infection systems) of selection for replication-competent virus. Ugandan subtypes A and D HIV-1 env sequences (115-A, 120-A, 89-D, 122-D, 126-D) were employed in all three assay systems. These subtypes co-circulate in East Africa and frequently recombine in this human population.
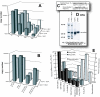
Results: Increased sequence identity between viruses or RNA templates resulted in increased recombination frequencies, with the exception of the 115-A virus or RNA template. Analyses of the recombination breakpoints and mechanistic studies revealed that the presence of a recombination hotspot in the C3/V4 env region, unique to 115-A as donor RNA, could account for the higher recombination frequencies with the 115-A virus/template. Single-cycle infections supported proportionally less recombination than the in vitro reverse transcription assay but both systems still had significantly higher recombination frequencies than observed in the multiple-cycle virus replication system. In the multiple cycle assay, increased replicative fitness of one HIV-1 over the other in a dual infection dramatically decreased recombination frequencies.
Conclusion: Sequence variation at specific sites between HIV-1 isolates can introduce unique recombination hotspots, which increase recombination frequencies and skew the general observation that decreased HIV-1 sequence identity reduces recombination rates. These findings also suggest that the majority of intra- or intersubtype A/D HIV-1 recombinants, generated with each round of infection, are not replication-competent and do not survive in the multiple-cycle system. Ability of one HIV-1 isolate to outgrow the other leads to reduced co-infections, heterozygous virus production, and recombination frequencies.
Figures



References
-
- Hu WS, Pathak VK, Temin HM. Role of reverse transcriptase in retroviral recombination. In: Skalka AM and Goff SP, editor. Reverse transcruptase. New York, Cold Spring Harbor Laboratory Press; 1993. pp. 251–274.
Publication types
MeSH terms
Substances
Grants and funding
- R21 AI049170/AI/NIAID NIH HHS/United States
- AI49170/AI/NIAID NIH HHS/United States
- R56 AI049170/AI/NIAID NIH HHS/United States
- AI25879/AI/NIAID NIH HHS/United States
- GM051140/GM/NIGMS NIH HHS/United States
- AI43645-02/AI/NIAID NIH HHS/United States
- AI57005/AI/NIAID NIH HHS/United States
- R29 GM051140/GM/NIGMS NIH HHS/United States
- P01 AI057005/AI/NIAID NIH HHS/United States
- R01 AI049170/AI/NIAID NIH HHS/United States
- R01 GM051140/GM/NIGMS NIH HHS/United States
- U01 AI025879/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

