Bursts of nonsynonymous substitutions in HIV-1 evolution reveal instances of positive selection at conservative protein sites
- PMID: 17164328
- PMCID: PMC1698441
- DOI: 10.1073/pnas.0609484103
Bursts of nonsynonymous substitutions in HIV-1 evolution reveal instances of positive selection at conservative protein sites
Abstract
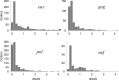
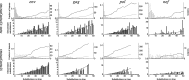
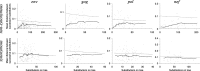
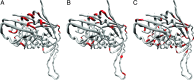
The fixation of a new allele can be driven by Darwinian positive selection or can be due to random genetic drift. Identifying instances of positive selection is a difficult task, because its impact is routinely obscured by the action of negative selection. The nature of the genetic code dictates that positive selection in favor of an amino acid replacement should often cause a burst of two or three nucleotide substitutions at a single codon site, because a large fraction of amino acid replacements cannot be achieved after just one nucleotide substitution. Here, we study pairs of successive nonsynonymous substitutions at one codon in the course of evolution of HIV-1 genes within HIV-1 populations inhabiting infected individuals. Such pairs are more numerous and more clumped than expected if different substitutions were independent and than what is observed for pairs of successive synonymous substitutions. Bursts of nonsynonymous substitutions in HIV-1 evolution cannot be explained by mutational biases and must, therefore, be due to positive selection. Both reversals, exact or imprecise, of fixed deleterious mutations and acquisitions of amino acids with new properties are responsible for the bursts. Temporal clumping is strongest at codon sites with a low overall rate of nonsynonymous evolution, implying that a substantial fraction of replacements of conservative amino acids are driven by positive selection. We identified many conservative sites of HIV-1 proteins that occasionally experience positive selection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Inferring natural selection operating on conservative and radical substitution at single amino acid sites.Genes Genet Syst. 2007 Aug;82(4):341-60. doi: 10.1266/ggs.82.341. Genes Genet Syst. 2007. PMID: 17895585
-
Positive selection at sites of multiple amino acid replacements since rat-mouse divergence.Nature. 2004 Jun 3;429(6991):558-62. doi: 10.1038/nature02601. Nature. 2004. PMID: 15175752
-
A method for detecting positive selection at single amino acid sites.Mol Biol Evol. 1999 Oct;16(10):1315-28. doi: 10.1093/oxfordjournals.molbev.a026042. Mol Biol Evol. 1999. PMID: 10563013
-
Extensive purifying selection acting on synonymous sites in HIV-1 Group M sequences.Virol J. 2008 Dec 23;5:160. doi: 10.1186/1743-422X-5-160. Virol J. 2008. PMID: 19105834 Free PMC article. Review.
-
Inference of selection from multiple species alignments.Curr Opin Genet Dev. 2002 Dec;12(6):688-94. doi: 10.1016/s0959-437x(02)00348-9. Curr Opin Genet Dev. 2002. PMID: 12433583 Review.
Cited by
-
Recombination rate and selection strength in HIV intra-patient evolution.PLoS Comput Biol. 2010 Jan 29;6(1):e1000660. doi: 10.1371/journal.pcbi.1000660. PLoS Comput Biol. 2010. PMID: 20126527 Free PMC article.
-
The evolutionary significance of certain amino acid substitutions and their consequences for HIV-1 immunogenicity toward HLA's A*0201 and B*27.Bioinformation. 2013 Mar 19;9(6):315-20. doi: 10.6026/97320630009315. Print 2013. Bioinformation. 2013. PMID: 23745018 Free PMC article.
-
Different Patterns of Codon Usage and Amino Acid Composition across Primate Lentiviruses.Viruses. 2023 Jul 20;15(7):1580. doi: 10.3390/v15071580. Viruses. 2023. PMID: 37515266 Free PMC article.
-
Prevalence of epistasis in the evolution of influenza A surface proteins.PLoS Genet. 2011 Feb;7(2):e1001301. doi: 10.1371/journal.pgen.1001301. Epub 2011 Feb 17. PLoS Genet. 2011. PMID: 21390205 Free PMC article.
-
Dinucleotide codon substitutions as a signature of diversifying selection in Mycobacterium tuberculosis.Front Mol Biosci. 2025 Jul 2;12:1511372. doi: 10.3389/fmolb.2025.1511372. eCollection 2025. Front Mol Biosci. 2025. PMID: 40671870 Free PMC article.
References
-
- Fisher RA. The Genetical Theory of Natural Selection. Oxford, UK: Clarendon; 1930.
-
- Williams GC. Adaptation and Natural Selection; A Critique of Some Current Evolutionary Thought. Princeton, NJ: Princeton Univ Press; 1966.
-
- Nielsen R. Annu Rev Genet. 2005;39:197–218. - PubMed
-
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, UK: Cambridge Univ Press; 1983.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

