An allosteric-feedback mechanism for protein-assisted group I intron splicing
- PMID: 17164477
- PMCID: PMC1781373
- DOI: 10.1261/rna.307907
An allosteric-feedback mechanism for protein-assisted group I intron splicing
Abstract
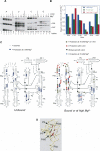
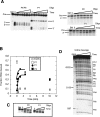
The I-AniI maturase facilitates self-splicing of a mitochondrial group I intron in Aspergillus nidulans. Binding occurs in at least two steps: first, a specific but labile encounter complex rapidly forms and then this intermediate is slowly resolved into a native, catalytically active RNA/protein complex. Here we probe the structure of the RNA throughout the assembly pathway. Although inherently unstable, the intron core, when bound by I-AniI, undergoes rapid folding to a near-native state in the encounter complex. The next transition includes the slow destabilization and docking into the core of the peripheral stacked helix that contains the 5' splice site. Mutational analyses confirm that both transitions are important for native complex formation. We propose that protein-driven destabilization and docking of the peripheral stacked helix lead to subtle changes in the I-AniI binding site that facilitate native complex formation. These results support an allosteric-feedback mechanism of RNA-protein recognition in which proteins engaged in an intermediate complex can influence RNA structure far from their binding sites. The linkage of these changes to stable binding ensures that the protein and RNA do not get sequestered in nonfunctional complexes.
Figures


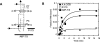
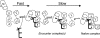
Similar articles
-
The maturase encoded by a group I intron from Aspergillus nidulans stabilizes RNA tertiary structure and promotes rapid splicing.J Mol Biol. 1999 Oct 8;292(5):987-1001. doi: 10.1006/jmbi.1999.3070. J Mol Biol. 1999. PMID: 10512698
-
A comprehensive characterization of a group IB intron and its encoded maturase reveals that protein-assisted splicing requires an almost intact intron RNA.J Mol Biol. 2001 May 11;308(4):609-22. doi: 10.1006/jmbi.2001.4609. J Mol Biol. 2001. PMID: 11350164
-
A novel mechanism for protein-assisted group I intron splicing.RNA. 2002 Apr;8(4):412-25. doi: 10.1017/s1355838202029321. RNA. 2002. PMID: 11991637 Free PMC article.
-
A C-terminal fragment of an intron-encoded maturase is sufficient for promoting group I intron splicing.RNA. 2005 Apr;11(4):437-46. doi: 10.1261/rna.7225205. RNA. 2005. PMID: 15769873 Free PMC article.
-
Pre-mRNA splicing in yeast.Trends Genet. 1991 Mar;7(3):79-85. doi: 10.1016/0168-9525(91)90276-V. Trends Genet. 1991. PMID: 2031287 Review.
Cited by
-
Mitochondrial genome of Phlebia radiata is the second largest (156 kbp) among fungi and features signs of genome flexibility and recent recombination events.PLoS One. 2014 May 13;9(5):e97141. doi: 10.1371/journal.pone.0097141. eCollection 2014. PLoS One. 2014. PMID: 24824642 Free PMC article.
-
Evidence that binding of C5 protein to P RNA enhances ribozyme catalysis by influencing active site metal ion affinity.RNA. 2007 Sep;13(9):1505-15. doi: 10.1261/rna.571007. Epub 2007 Jul 25. RNA. 2007. PMID: 17652407 Free PMC article.
-
Conformational heterogeneity and the determinants of tertiary stabilization in the hammerhead ribozyme from Dolichopoda cave crickets.RNA Biol. 2011 Sep-Oct;8(5):893-903. doi: 10.4161/rna.8.5.16036. Epub 2011 Jun 29. RNA Biol. 2011. PMID: 21712651 Free PMC article.
-
A DExH/D-box protein coordinates the two steps of splicing in a group I intron.J Mol Biol. 2008 Nov 14;383(3):667-82. doi: 10.1016/j.jmb.2008.08.070. Epub 2008 Sep 4. J Mol Biol. 2008. PMID: 18789947 Free PMC article.
-
Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation.Nucleic Acids Res. 2009 Feb;37(3):877-90. doi: 10.1093/nar/gkn1007. Epub 2008 Dec 22. Nucleic Acids Res. 2009. PMID: 19103658 Free PMC article.
References
-
- Allain, F.H., Gubser, C.C., Howe, P.W., Nagai, K., Neuhaus, D., Varani, G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature. 1996;380:646–650. - PubMed
-
- Avis, J.M., Allain, F.H., Howe, P.W., Varani, G., Nagai, K., Neuhaus, D. Solution structure of the N-terminal RNP domain of U1A protein: The role of C-terminal residues in structure stability and RNA binding. J. Mol. Biol. 1996;257:398–411. - PubMed
-
- Bartley, L.E., Zhuang, X., Das, R., Chu, S., Herschlag, D. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J. Mol. Biol. 2003;328:1011–1026. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
