Crystallographic refinement of ligand complexes
- PMID: 17164531
- PMCID: PMC2483469
- DOI: 10.1107/S0907444906022657
Crystallographic refinement of ligand complexes
Abstract
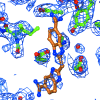
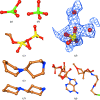
Model building and refinement of complexes between biomacromolecules and small molecules requires sensible starting coordinates as well as the specification of restraint sets for all but the most common non-macromolecular entities. Here, it is described why this is necessary, how it can be accomplished and what pitfalls need to be avoided in order to produce chemically plausible models of the low-molecular-weight entities. A number of programs, servers, databases and other resources that can be of assistance in the process are also discussed.
Figures
References
-
- Aalten, D. M. F. van, Bywater, R., Findlay, J. B. C., Hendlich, M., Hooft, R. W. W. & Vriend, G. (1996). J. Comput. Aided Mol. Des.10, 255–262. - PubMed
-
- Berman, H., Henrick, K. & Nakamura, H. (2003). Nature Struct. Biol.10, 980. - PubMed
-
- Bohne, A., Lang, E. & von der Lieth, C. W. (1999). Bioinformatics, 15, 767–768. - PubMed
-
- Boström, J. (2001). J. Comput. Aided Mol. Des.15, 1137–1152. - PubMed
-
- Brünger, A. T. (1992). X-PLOR. Version 3.1. A System for X-ray Crystallography and NMR. Yale University, Connecticut, USA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

