Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast
- PMID: 17166056
- PMCID: PMC1657058
- DOI: 10.1371/journal.pgen.0020203
Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast
Abstract
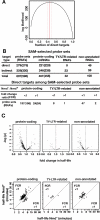
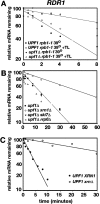
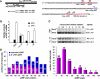
Nonsense-mediated mRNA decay (NMD) is a eukaryotic mechanism of RNA surveillance that selectively eliminates aberrant transcripts coding for potentially deleterious proteins. NMD also functions in the normal repertoire of gene expression. In Saccharomyces cerevisiae, hundreds of endogenous RNA Polymerase II transcripts achieve steady-state levels that depend on NMD. For some, the decay rate is directly influenced by NMD (direct targets). For others, abundance is NMD-sensitive but without any effect on the decay rate (indirect targets). To distinguish between direct and indirect targets, total RNA from wild-type (Nmd(+)) and mutant (Nmd(-)) strains was probed with high-density arrays across a 1-h time window following transcription inhibition. Statistical models were developed to describe the kinetics of RNA decay. 45% +/- 5% of RNAs targeted by NMD were predicted to be direct targets with altered decay rates in Nmd(-) strains. Parallel experiments using conventional methods were conducted to empirically test predictions from the global experiment. The results show that the global assay reliably distinguished direct versus indirect targets. Different types of targets were investigated, including transcripts containing adjacent, disabled open reading frames, upstream open reading frames, and those prone to out-of-frame initiation of translation. Known targeting mechanisms fail to account for all of the direct targets of NMD, suggesting that additional targeting mechanisms remain to be elucidated. 30% of the protein-coding targets of NMD fell into two broadly defined functional themes: those affecting chromosome structure and behavior and those affecting cell surface dynamics. Overall, the results provide a preview for how expression profiles in multi-cellular eukaryotes might be impacted by NMD. Furthermore, the methods for analyzing decay rates on a global scale offer a blueprint for new ways to study mRNA decay pathways in any organism where cultured cell lines are available.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures






References
-
- Culbertson MR, Leeds P. Looking at mRNA decay pathways through the window of molecular evolution. Curr Op Gen Dev. 2003;13:207–214. - PubMed
-
- Lynch M, Richardson AO. The evolution of spliceosomal introns. Curr Op Gen Dev. 2002;12:701–710. - PubMed
-
- Culbertson M. RNA surveillance: Unforeseen consequences for gene expression, inherited genetic disorders, and cancer. Trends Genet. 1999;15:74–80. - PubMed
-
- He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

