Microsatellite diversity and broad scale geographic structure in a model legume: building a set of nested core collection for studying naturally occurring variation in Medicago truncatula
- PMID: 17166278
- PMCID: PMC1762007
- DOI: 10.1186/1471-2229-6-28
Microsatellite diversity and broad scale geographic structure in a model legume: building a set of nested core collection for studying naturally occurring variation in Medicago truncatula
Abstract
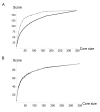
Background: Exploiting genetic diversity requires previous knowledge of the extent and structure of the variation occurring in a species. Such knowledge can in turn be used to build a core-collection, i.e. a subset of accessions that aim at representing the genetic diversity of this species with a minimum of repetitiveness. We investigate the patterns of genetic diversity and population structure in a collection of 346 inbred lines representing the breadth of naturally occurring diversity in the Legume plant model Medicago truncatula using 13 microsatellite loci distributed throughout the genome.
Results: We confirm the uniqueness of all these genotypes and reveal a large amount of genetic diversity and allelic variation within this autogamous species. Spatial genetic correlation was found only for individuals originating from the same population and between neighbouring populations. Using a model-based clustering algorithm, we identified four main genetic clusters in the set of individuals analyzed. This stratification matches broad geographic regions. We also identified a set of "admixed" individuals that do not fit with this population structure scheme.
Conclusion: The stratification inferred is discussed considering potential historical events like expansion, refuge history and admixture between neighbouring groups. Information on the allelic richness and the inferred population structure are used to build a nested core-collection. The set of inbred lines and the core collections are publicly available and will help coordinating efforts for the study of naturally occurring variation in the growing Medicago truncatula community.
Figures


Similar articles
-
Quality of core collections for effective utilisation of genetic resources review, discussion and interpretation.Theor Appl Genet. 2013 Feb;126(2):289-305. doi: 10.1007/s00122-012-1971-y. Epub 2012 Sep 15. Theor Appl Genet. 2013. PMID: 22983567 Free PMC article. Review.
-
How multilocus genotypic pattern helps to understand the history of selfing populations: a case study in Medicago truncatula.Heredity (Edinb). 2008 May;100(5):517-25. doi: 10.1038/hdy.2008.5. Epub 2008 Feb 20. Heredity (Edinb). 2008. PMID: 18285811
-
The use of neutral and non-neutral SSRs to analyse the genetic structure of a Tunisian collection of Medicago truncatula lines and to reveal associations with eco-environmental variables.Genetica. 2009 Apr;135(3):391-402. doi: 10.1007/s10709-008-9285-3. Epub 2008 Aug 14. Genetica. 2009. PMID: 18704697
-
SSR analysis of the Medicago truncatula SARDI core collection reveals substantial diversity and unusual genotype dispersal throughout the Mediterranean basin.Theor Appl Genet. 2006 Mar;112(5):977-83. doi: 10.1007/s00122-005-0202-1. Epub 2006 Jan 10. Theor Appl Genet. 2006. PMID: 16402186
-
Single-nucleotide polymorphism discovery and diversity in the model legume Medicago truncatula.Mol Ecol Resour. 2013 Jan;13(1):84-95. doi: 10.1111/1755-0998.12021. Epub 2012 Sep 27. Mol Ecol Resour. 2013. PMID: 23017123
Cited by
-
Molecular adaptation in flowering and symbiotic recognition pathways: insights from patterns of polymorphism in the legume Medicago truncatula.BMC Evol Biol. 2011 Aug 1;11:229. doi: 10.1186/1471-2148-11-229. BMC Evol Biol. 2011. PMID: 21806823 Free PMC article.
-
Quality of core collections for effective utilisation of genetic resources review, discussion and interpretation.Theor Appl Genet. 2013 Feb;126(2):289-305. doi: 10.1007/s00122-012-1971-y. Epub 2012 Sep 15. Theor Appl Genet. 2013. PMID: 22983567 Free PMC article. Review.
-
Investigation of the demographic and selective forces shaping the nucleotide diversity of genes involved in nod factor signaling in Medicago truncatula.Genetics. 2007 Dec;177(4):2123-33. doi: 10.1534/genetics.107.076943. Genetics. 2007. PMID: 18073426 Free PMC article.
-
Genetic diversity assessment of sesame core collection in China by phenotype and molecular markers and extraction of a mini-core collection.BMC Genet. 2012 Nov 15;13:102. doi: 10.1186/1471-2156-13-102. BMC Genet. 2012. PMID: 23153260 Free PMC article.
-
Genetic Diversity and Population Structure Analysis of Castanopsis hystrix and Construction of a Core Collection Using Phenotypic Traits and Molecular Markers.Genes (Basel). 2022 Dec 16;13(12):2383. doi: 10.3390/genes13122383. Genes (Basel). 2022. PMID: 36553650 Free PMC article.
References
-
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lange BM, Moughamer T, Xia Y, Budworth P, Zhong J, Miguel T, Paszkowski U, Zhang S, Colbert M, Sun WL, Chen L, Cooper B, Park S, Wood TC, Mao L, Quail P, Wing R, Dean R, Yu Y, Zharkikh A, Shen R, Sahasrabudhe S, Thomas A, Cannings R, Gutin A, Pruss D, Reid J, Tavtigian S, Mitchell J, Eldredge G, Scholl T, Miller RM, Bhatnagar S, Adey N, Rubano T, Tusneem N, Robinson R, Feldhaus J, Macalma T, Oliphant A, Briggs S. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. - DOI - PubMed
-
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G, Huang X, Li W, Li J, Liu Z, Li L, Liu J, Qi Q, Liu J, Li L, Li T, Wang X, Lu H, Wu T, Zhu M, Ni P, Han H, Dong W, Ren X, Feng X, Cui P, Li X, Wang H, Xu X, Zhai W, Xu Z, Zhang J, He S, Zhang J, Xu J, Zhang K, Zheng X, Dong J, Zeng W, Tao L, Ye J, Tan J, Ren X, Chen X, He J, Liu D, Tian W, Tian C, Xia H, Bao Q, Li G, Gao H, Cao T, Wang J, Zhao W, Li P, Chen W, Wang X, Zhang Y, Hu J, Wang J, Liu S, Yang J, Zhang G, Xiong Y, Li Z, Mao L, Zhou C, Zhu Z, Chen R, Hao B, Zheng W, Chen S, Guo W, Li G, Liu S, Tao M, Wang J, Zhu L, Yuan L, Yang H. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. - DOI - PubMed
-
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, Jakobsson M, Kim S, Morozov Y, Padhukasahasram B, Plagnol V, Rosenberg NA, Shah C, Wall JD, Wang J, Zhao K, Kalbfleisch T, Schulz V, Kreitman M, Bergelson J. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

