Directionality of point mutation and 5-methylcytosine deamination rates in the chimpanzee genome
- PMID: 17166280
- PMCID: PMC1764022
- DOI: 10.1186/1471-2164-7-316
Directionality of point mutation and 5-methylcytosine deamination rates in the chimpanzee genome
Abstract
Background: The pattern of point mutation is important for studying mutational mechanisms, genome evolution, and diseases. Previous studies of mutation direction were largely based on substitution data from a limited number of loci. To date, there is no genome-wide analysis of mutation direction or methylation-dependent transition rates in the chimpanzee or its categorized genomic regions.
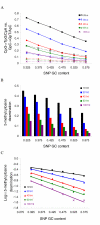
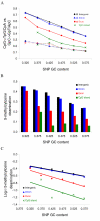
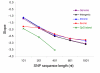
Results: In this study, we performed a detailed examination of mutation direction in the chimpanzee genome and its categorized genomic regions using 588,918 SNPs whose ancestral alleles could be inferred by mapping them to human genome sequences. The C-->T (G-->A) changes occurred most frequently in the chimpanzee genome. Each type of transition occurred approximately four times more frequently than each type of transversion. Notably, the frequency of C-->T (G-->A) was the highest in exons among the genomic categories regardless of whether we calculated directly, normalized with the nucleotide content, or removed the SNPs involved in the CpG effect. Moreover, the directionality of the point mutation in exons and CpG islands were opposite relative to their corresponding intergenic regions, indicating that different forces govern the nucleotide changes. Our analysis suggests that the GC content is not in equilibrium in the chimpanzee genome. Further quantitative analysis revealed that the 5-methylcytosine deamination rates at CpG sites were highly dependent on the local GC content and the lengths of SNP flanking sequences and varied among categorized genomic regions.
Conclusion: We present the first mutational spectrum, estimated by three different approaches, in the chimpanzee genome. Our results provide detailed information on recent nucleotide changes and methylation-dependent transition rates in the chimpanzee genome after its split from the human. These results have important implications for understanding genome composition evolution, mechanisms of point mutation, and other genetic factors such as selection, biased codon usage, biased gene conversion, and recombination.
Figures
Similar articles
-
Mutational spectrum in the recent human genome inferred by single nucleotide polymorphisms.Genomics. 2006 Nov;88(5):527-34. doi: 10.1016/j.ygeno.2006.06.003. Epub 2006 Jul 24. Genomics. 2006. PMID: 16860534
-
CpG mutation rates in the human genome are highly dependent on local GC content.Mol Biol Evol. 2005 Mar;22(3):650-8. doi: 10.1093/molbev/msi043. Epub 2004 Nov 10. Mol Biol Evol. 2005. PMID: 15537806
-
Strand bias in complementary single-nucleotide polymorphisms of transcribed human sequences: evidence for functional effects of synonymous polymorphisms.BMC Genomics. 2006 Aug 17;7:213. doi: 10.1186/1471-2164-7-213. BMC Genomics. 2006. PMID: 16916449 Free PMC article.
-
[Chimpanzee genome sequencing and comparative analysis with the human genome].Tanpakushitsu Kakusan Koso. 2006 Feb;51(2):178-87. Tanpakushitsu Kakusan Koso. 2006. PMID: 16457209 Review. Japanese. No abstract available.
-
Understanding the recent evolution of the human genome: insights from human-chimpanzee genome comparisons.Hum Mutat. 2007 Feb;28(2):99-130. doi: 10.1002/humu.20420. Hum Mutat. 2007. PMID: 17024666 Review.
Cited by
-
Accumulation of GC donor splice signals in mammals.Biol Direct. 2008 Jul 9;3:30. doi: 10.1186/1745-6150-3-30. Biol Direct. 2008. PMID: 18613975 Free PMC article.
-
Brain aging and Alzheimer's disease, a perspective from non-human primates.Aging (Albany NY). 2024 Oct 29;16(20):13145-13171. doi: 10.18632/aging.206143. Epub 2024 Oct 29. Aging (Albany NY). 2024. PMID: 39475348 Free PMC article. Review.
-
Human-specific CpG "beacons" identify loci associated with human-specific traits and disease.Epigenetics. 2012 Oct;7(10):1188-99. doi: 10.4161/epi.22127. Epub 2012 Sep 11. Epigenetics. 2012. PMID: 22968434 Free PMC article.
-
Comparative analysis reveals epigenomic evolution related to species traits and genomic imprinting in mammals.Innovation (Camb). 2023 Apr 28;4(3):100434. doi: 10.1016/j.xinn.2023.100434. eCollection 2023 May 15. Innovation (Camb). 2023. PMID: 37215528 Free PMC article.
-
Sex and parasites: genomic and transcriptomic analysis of Microbotryum lychnidis-dioicae, the biotrophic and plant-castrating anther smut fungus.BMC Genomics. 2015 Jun 16;16(1):461. doi: 10.1186/s12864-015-1660-8. BMC Genomics. 2015. PMID: 26076695 Free PMC article.
References
-
- Watanabe H, Fujiyama A, Hattori M, Taylor TD, Toyoda A, Kuroki Y, Noguchi H, BenKahla A, Lehrach H, Sudbrak R, Kube M, Taenzer S, Galgoczy P, Platzer M, Scharfe M, Nordsiek G, Blocker H, Hellmann I, Khaitovich P, Paabo S, Reinhardt R, Zheng HJ, Zhang XL, Zhu GF, Wang BF, Fu G, Ren SX, Zhao GP, Chen Z, Lee YS, Cheong JE, Choi SH, Wu KM, Liu TT, Hsiao KJ, Tsai SF, Kim CG, S OO, Kitano T, Kohara Y, Saitou N, Park HS, Wang SY, Yaspo ML, Sakaki Y. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature. 2004;429:382–388. doi: 10.1038/nature02564. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

