Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics
- PMID: 17166288
- PMCID: PMC1762009
- DOI: 10.1186/1472-6807-6-26
Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics
Abstract
Background: The histone H3/H4 chaperone Asf1 (anti-silencing function 1) is required for the establishment and maintenance of proper chromatin structure, as well as for genome stability in eukaryotes. Asf1 participates in both DNA replication-coupled (RC) and replication-independent (RI) histone deposition reactions in vitro and interacts with complexes responsible for both pathways in vivo. Asf1 is known to directly bind histone H3, however, high-resolution structural information about the geometry of this interaction was previously unknown.
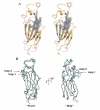
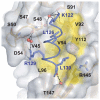
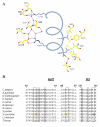
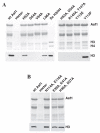
Results: Here we report the structure of a histone/histone chaperone interaction. We have solved the 2.2 A crystal structure of the conserved N-terminal immunoglobulin fold domain of yeast Asf1 (residues 2-155) bound to the C-terminal helix of yeast histone H3 (residues 121-134). The structure defines a histone-binding patch on Asf1 consisting of both conserved and yeast-specific residues; mutation of these residues abrogates H3/H4 binding affinity. The geometry of the interaction indicates that Asf1 binds to histones H3/H4 in a manner that likely blocks sterically the H3/H3 interface of the nucleosomal four-helix bundle.
Conclusion: These data clarify how Asf1 regulates histone stoichiometry to modulate epigenetic inheritance. The structure further suggests a physical model in which Asf1 contributes to interpretation of a "histone H3 barcode" for sorting H3 isoforms into different deposition pathways.
Figures

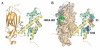
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

