Three-dimensional structure determined for a subunit of human tRNA splicing endonuclease (Sen15) reveals a novel dimeric fold
- PMID: 17166513
- PMCID: PMC1865571
- DOI: 10.1016/j.jmb.2006.11.024
Three-dimensional structure determined for a subunit of human tRNA splicing endonuclease (Sen15) reveals a novel dimeric fold
Abstract
Splicing of eukaryal intron-containing tRNAs requires the action of the heterotetrameric splicing endonuclease, which is composed of two catalytic subunits, Sen34 and Sen2, and two structural subunits, Sen15 and Sen54. Here we report the solution structure of the human tRNA splicing endonuclease subunit HsSen15. To facilitate the structure determination, we removed the disordered 35 N-terminal and 14 C-terminal residues of the full-length protein to produce HsSen15(36-157). The structure of HsSen15(36-157), the first for a subunit of a eukaryal splicing endonuclease, revealed that the protein possesses a novel homodimeric fold. Each monomer consists of three alpha-helices and a mixed antiparallel/parallel beta-sheet, arranged in a topology similar to that of the C-terminal domain of Methanocaldococcus jannaschii endonuclease. The dimeric interface is dominated by a beta-barrel structure, formed by face-to-face packing of two, three-stranded beta-sheets. Each of the beta-sheets results from reciprocal parallel pairing of one beta-strand from one subunit with two other beta-strands from the symmetric subunit. The structural model provides insights into the functional assembly of the human tRNA splicing endonuclease.
Figures


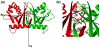

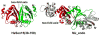

Similar articles
-
Crystal structure of a dimeric archaeal splicing endonuclease.J Mol Biol. 2000 Sep 22;302(3):639-48. doi: 10.1006/jmbi.2000.3941. J Mol Biol. 2000. PMID: 10986124
-
Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site.Nature. 2006 May 18;441(7091):375-7. doi: 10.1038/nature04741. Nature. 2006. PMID: 16710424
-
A novel three-unit tRNA splicing endonuclease found in ultrasmall Archaea possesses broad substrate specificity.Nucleic Acids Res. 2011 Dec;39(22):9695-704. doi: 10.1093/nar/gkr692. Epub 2011 Aug 31. Nucleic Acids Res. 2011. PMID: 21880595 Free PMC article.
-
RNA-splicing endonuclease structure and function.Cell Mol Life Sci. 2008 Apr;65(7-8):1176-85. doi: 10.1007/s00018-008-7393-y. Cell Mol Life Sci. 2008. PMID: 18217203 Free PMC article. Review.
-
Evolutionary markers in the (beta/alpha)8-barrel fold.Curr Opin Chem Biol. 2003 Dec;7(6):694-701. doi: 10.1016/j.cbpa.2003.10.004. Curr Opin Chem Biol. 2003. PMID: 14644177 Review.
Cited by
-
The dawn of dominance by the mature domain in tRNA splicing.Proc Natl Acad Sci U S A. 2007 Jul 24;104(30):12300-5. doi: 10.1073/pnas.0705537104. Epub 2007 Jul 16. Proc Natl Acad Sci U S A. 2007. PMID: 17636125 Free PMC article.
-
Recent Insights Into the Structure, Function, and Evolution of the RNA-Splicing Endonucleases.Front Genet. 2019 Feb 12;10:103. doi: 10.3389/fgene.2019.00103. eCollection 2019. Front Genet. 2019. PMID: 30809252 Free PMC article. Review.
-
Structural basis for RNA recognition by a type II poly(A)-binding protein.Proc Natl Acad Sci U S A. 2008 Oct 7;105(40):15317-22. doi: 10.1073/pnas.0801274105. Epub 2008 Sep 29. Proc Natl Acad Sci U S A. 2008. PMID: 18824697 Free PMC article.
-
Reconstitution of the human tRNA splicing endonuclease complex: insight into the regulation of pre-tRNA cleavage.Nucleic Acids Res. 2020 Aug 20;48(14):7609-7622. doi: 10.1093/nar/gkaa438. Nucleic Acids Res. 2020. PMID: 32476018 Free PMC article.
-
Mediation of the Same Epigenetic and Transcriptional Effect by Independent Osteoarthritis Risk-Conferring Alleles on a Shared Target Gene, COLGALT2.Arthritis Rheumatol. 2023 Jun;75(6):910-922. doi: 10.1002/art.42427. Epub 2023 Apr 11. Arthritis Rheumatol. 2023. PMID: 36538011 Free PMC article.
References
-
- Cech T. In: RNA World. Gesteland RF, Atkins JF, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1993. pp. 239–270.
-
- Peebles CL, Gegenheimer P, Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell. 1983;32:525–536. - PubMed
-
- Thompson LD, Daniels CJ. Recognition of exon-intron boundaries by the Halobacterium volcanii tRNA intron endonuclease. J Biol Chem. 1990;265:18104–18111. - PubMed
-
- Trotta CR, Miao F, Arn EA, Stevens SW, Ho CK, Rauhut R, et al. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell. 1997;89:849–858. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

