HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry
- PMID: 17166540
- PMCID: PMC1892195
- DOI: 10.1016/j.virol.2006.11.004
HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry
Abstract
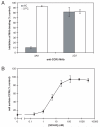
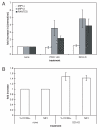
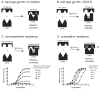
Human immunodeficiency virus type 1 (HIV-1) infection can be inhibited by small molecules that target the CCR5 coreceptor. Here, we describe some properties of clonal viruses resistant to one such inhibitor, SCH-D, using both chimeric, infectious molecular clones and Env-pseudotypes. Studies using combinations of CCR5 ligands, including small molecule inhibitors, monoclonal antibodies (MAbs) and chemokine derivatives such as PSC-RANTES, show that the fully SCH-D-resistant viruses enter target cells by using the SCH-D-bound form of CCR5. However, the way resistance to SCH-D and other small molecule CCR5 inhibitors is manifested depends on the target cell and the nature of the assay (single- vs. multi-cycle). In multi-cycle assays using primary lymphocytes, SCH-D does not inhibit resistant molecular clones, and it can even enhance their infectivity modestly. In contrast, the same viruses (as Env-pseudotypes) are significantly inhibited by SCH-D in single-cycle entry assays using U87-CD4/CCR5 cells, resistance being manifested by incomplete inhibition at high SCH-D concentrations. When a single-cycle, Env-pseudotype entry assay was performed using either U87-CD4/CCR5 cells or PBMC under comparable conditions, entry was inhibited by up to 88% in the former cells but by only 28% in the PBMC. Hence, there are both cell- and assay-dependent influences on how resistance is manifested. We also take this opportunity to correct our previous report that SCH-D-resistant isolates are also substantially cross-resistant to PSC-RANTES [Marozsan, A.J., Kuhmann, S.E., Morgan, T., Herrera, C., Rivera-Troche, E., Xu, S., Baroudy, B.M., Strizki, J., Moore, J.P., 2005. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D). Virology 338 (1), 182-199]. A substantial element of this resistance was attributable to the unappreciated carry-over of SCH-D from the selection cultures into analytical assays.
Figures




References
-
- Billick E, Seibert C, Pugach P, Ketas T, Trkola A, Endres MJ, Murgolo NJ, Coates E, Reyes GR, Baroudy BM, Sakmar TP, Moore JP, Kuhmann SE. The differential sensitivity of human and rhesus macaque CCR5 to small-molecule inhibitors of human immunodeficiency virus type 1 entry is explained by a single amino acid difference and suggests a mechanism of action for these inhibitors. J. Virol. 2004;78(8):4134–4144. - PMC - PubMed
-
- Coakley E, Petropoulos CJ, Whitcomb JM. Assessing chemokine co-receptor usage in HIV. Curr. Opin. Infect. Dis. 2005;18(1):9–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

