Evolutionary history and global spread of the emerging g12 human rotaviruses
- PMID: 17166908
- PMCID: PMC1865926
- DOI: 10.1128/JVI.01622-06
Evolutionary history and global spread of the emerging g12 human rotaviruses
Abstract
G12 rotaviruses were first detected in diarrheic children in the Philippines in 1987, but no further cases were reported until 1998. However, G12 rotaviruses have been detected all over the world in recent years. Here, we report the worldwide variations of G12 rotaviruses to investigate the evolutionary mechanisms by which they managed to spread globally in a short period of time. We sequenced the complete genomes (11 segments) of nine G12 rotaviruses isolated in Bangladesh, Belgium, Thailand, and the Philippines and compared them with the genomes of other rotavirus strains. Our genetic analyses revealed that after introduction of the VP7 gene of the rare G12 genotype into more common local strains through reassortment, a vast genetic diversity was generated and several new variants with distinct gene constellations emerged. These reassortment events most likely took place in Southeast Asian countries and spread to other parts of the world. The acquirement of gene segments from human-adapted rotaviruses might allow G12 to better propagate in humans and hence to develop into an important emerging human pathogen.
Figures
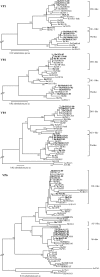
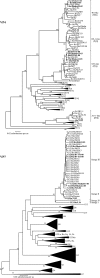
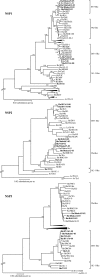
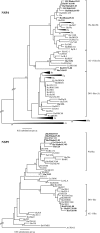

References
-
- Alfieri, A. A., J. P. Leite, O. Nakagomi, E. Kaga, P. A. Woods, R. I. Glass, and J. R. Gentsch. 1996. Characterization of human rotavirus genotype P[8]G5 from Brazil by probe-hybridization and sequence. Arch. Virol. 141:2353-2364. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Castello, A. A., M. L. Arvay, R. I. Glass, and J. Gentsch. 2004. Rotavirus strain surveillance in Latin America: a review of the last nine years. Pediatr. Infect. Dis. J. 23:S168-S172. - PubMed
-
- Castello, A. A., M. H. Arguelles, R. P. Rota, A. Olthoff, B. Jiang, R. I. Glass, J. R. Gentsch, and G. Glikmann. 2006. Molecular epidemiology of group A rotavirus diarrhea among children in Buenos Aires, Argentina, from 1999 to 2003 and emergence of the infrequent genotype G12. J. Clin. Microbiol. 44:2046-2050. - PMC - PubMed
-
- Cunliffe, N. A., J. S. Bresee, J. R. Gentsch, R. I. Glass, and C. A. Hart. 2002. The expanding diversity of rotaviruses. Lancet 359:640-642. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

