Regulation of methanol utilisation pathway genes in yeasts
- PMID: 17169150
- PMCID: PMC1781073
- DOI: 10.1186/1475-2859-5-39
Regulation of methanol utilisation pathway genes in yeasts
Abstract
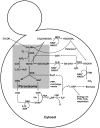
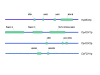
Methylotrophic yeasts such as Candida boidinii, Hansenula polymorpha, Pichia methanolica and Pichia pastoris are an emerging group of eukaryotic hosts for recombinant protein production with an ever increasing number of applications during the last 30 years. Their applications are linked to the use of strong methanol-inducible promoters derived from genes of the methanol utilisation pathway. These promoters are tightly regulated, highly repressed in presence of non-limiting concentrations of glucose in the medium and strongly induced if methanol is used as carbon source. Several factors involved in this tight control and their regulatory effects have been described so far. This review summarises available data about the regulation of promoters from methanol utilisation pathway genes. Furthermore, the role of cis and trans acting factors (e.g. transcription factors, glucose processing enzymes) in the expression of methanol utilisation pathway genes is reviewed both in the context of the native cell environment as well as in heterologous hosts.
Figures
References
-
- Wolf K. Nonconventional yeasts in biotechnology. Berlin, Heidelberg, New York, Springer; 1996.
-
- Buckholz RG, Gleeson MA. Yeast systems for the commercial production of heterologous proteins. Biotechnology (N Y) 1991;9:1067–1072. - PubMed
-
- Sudbery PE. The expression of recombinant proteins in yeasts. Curr Opin Biotechnol. 1996;7:517–524. - PubMed
-
- Dominguez A, Ferminan E, Sanchez M, Gonzalez FJ, Perez-Campo FM, Garcia S, Herrero AB, San Vicente A, Cabello J, Prado M, Iglesias FJ, Choupina A, Burguillo FJ, Fernandez-Lago L, Lopez MC. Non-conventional yeasts as hosts for heterologous protein production. Int Microbiol. 1998;1:131–142. - PubMed
-
- Müller S, Sandal T, Kamp-Hansen P, Dalboge H. Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast. 1998;14:1267–1283. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

