Crystal structure of human cyclin K, a positive regulator of cyclin-dependent kinase 9
- PMID: 17169370
- PMCID: PMC1852425
- DOI: 10.1016/j.jmb.2006.11.057
Crystal structure of human cyclin K, a positive regulator of cyclin-dependent kinase 9
Abstract
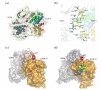
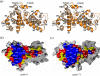
Cyclin K and the closely related cyclins T1, T2a, and T2b interact with cyclin-dependent kinase 9 (CDK9) forming multiple nuclear complexes, referred to collectively as positive transcription elongation factor b (P-TEFb). Through phosphorylation of the C-terminal domain of the RNA polymerase II largest subunit, distinct P-TEFb species regulate the transcriptional elongation of specific genes that play central roles in human physiology and disease development, including cardiac hypertrophy and human immunodeficiency virus-1 pathogenesis. We have determined the crystal structure of human cyclin K (residues 11-267) at 1.5 A resolution, which represents the first atomic structure of a P-TEFb subunit. The cyclin K fold comprises two typical cyclin boxes with two short helices preceding the N-terminal box. A prominent feature of cyclin K is an additional helix (H4a) in the first cyclin box that obstructs the binding pocket for the cell-cycle inhibitor p27(Kip1). Modeling of CDK9 bound to cyclin K provides insights into the structural determinants underlying the formation and regulation of this complex. A homology model of human cyclin T1 generated using the cyclin K structure as a template reveals that the two proteins have similar structures, as expected from their high level of sequence identity. Nevertheless, their CDK9-interacting surfaces display significant structural differences, which could potentially be exploited for the design of cyclin-targeted inhibitors of the CDK9-cyclin K and CDK9-cyclin T1 complexes.
Figures


References
-
- Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. - PubMed
-
- Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 2005;30:630–641. - PubMed
-
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. - PubMed
-
- Russo AA, Jeffrey PD, Pavletich NP. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nature Struct. Biol. 1996;3:696–700. - PubMed
-
- Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nature Cell Biol. 1999;1:438–443. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- DK062162/DK/NIDDK NIH HHS/United States
- R01 AG021964-03/AG/NIA NIH HHS/United States
- R01 DK062162-03/DK/NIDDK NIH HHS/United States
- R01 GM065520-02/GM/NIGMS NIH HHS/United States
- R01 GM065520/GM/NIGMS NIH HHS/United States
- R01 AG021964-04/AG/NIA NIH HHS/United States
- R01 GM065520-03/GM/NIGMS NIH HHS/United States
- AG021964/AG/NIA NIH HHS/United States
- R01 DK062162/DK/NIDDK NIH HHS/United States
- R01 GM065520-04/GM/NIGMS NIH HHS/United States
- R01 DK062162-04/DK/NIDDK NIH HHS/United States
- R01 AG021964/AG/NIA NIH HHS/United States
- GM065520/GM/NIGMS NIH HHS/United States
- R01 GM065520-01/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

