Selenium derivatization of nucleic acids for crystallography
- PMID: 17169989
- PMCID: PMC1802610
- DOI: 10.1093/nar/gkl1070
Selenium derivatization of nucleic acids for crystallography
Abstract
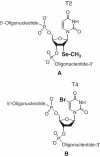
The high-resolution structure of the DNA (5'-GTGTACA-C-3') with the selenium derivatization at the 2'-position of T2 was determined via MAD and SAD phasing. The selenium-derivatized structure (1.28 A resolution) with the 2'-Se modification in the minor groove is isomorphorous to the native structure (2.0 A). To directly compare with the conventional bromine derivatization, we incorporated bromine into the 5-postion of T4, determined the bromine-derivatized DNA structure at 1.5 A resolution, and found that the local backbone torsion angles and solvent hydration patterns were altered in the structure with the Br incorporation in the major groove. Furthermore, while the native and Br-derivatized DNAs needed over a week to form reasonable-size crystals, we observed that the Se-derivatized DNAs grew crystals overnight with high-diffraction quality, suggesting that the Se derivatization facilitated the crystal formation. In addition, the Se-derivatized DNA sequences crystallized under a broader range of buffer conditions, and generally had a faster crystal growth rate. Our experimental results indicate that the selenium derivatization of DNAs may facilitate the determination of nucleic acid X-ray crystal structures in phasing and high-quality crystal growth. In addition, our results suggest that the Se derivatization can be an alternative to the conventional Br derivatization.
Figures



References
-
- MacRae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. - PubMed
-
- Jacobs S.A., Podell E.R., Cech T.R. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nature Struct. Mol. Biol. 2006;13:218–225. - PubMed
-
- Adams P.L., Stahley M.R., Kosek A.B., Wang J., Strobel S.A. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. - PubMed
-
- Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. - PubMed
-
- Peersen O.B., Ruggles J.A., Schultz S.C. Dimeric structure of the Oxytricha nova telomere end-binding protein alpha-subunit bound to ssDNA. Nature Struct. Biol. 2002;9:182–187. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

