Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes
- PMID: 17170000
- PMCID: PMC1802598
- DOI: 10.1093/nar/gkl1050
Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes
Abstract
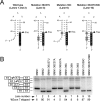
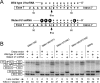
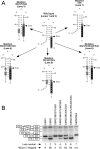
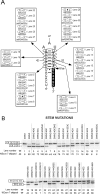
Humans have two nearly identical copies of the survival motor neuron (SMN ) gene, SMN1 and SMN2. Homozygous loss of SMN1 causes spinal muscular atrophy (SMA). SMN2 is unable to prevent the disease due to skipping of exon 7. Using a systematic approach of in vivo selection, we have previously demonstrated that a weak 5' splice site (ss) serves as the major cause of skipping of SMN2 exon 7. Here we show the inhibitory impact of RNA structure on the weak 5' ss of exon 7. We call this structure terminal stem-loop 2 (TSL2). Confirming the inhibitory nature of TSL2, point mutations that destabilize TSL2 promote exon 7 inclusion in SMN2, whereas strengthening of TSL2 promotes exon 7 skipping even in SMN1. We also demonstrate that TSL2 negatively affects the recruitment of U1snRNP at the 5' ss of exon 7. Using enzymatic structure probing, we confirm that the sequence at the junction of exon 7/intron 7 folds into TSL2 and show that mutations in TSL2 cause predicted structural changes in this region. Our findings reveal for the first time the critical role of RNA structure in regulation of alternative splicing of human SMN.
Figures



References
-
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. - PubMed
-
- Rothrock C., Cannon B., Hahm B., Lynch K.W. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol. Cell. 2003;12:1317–1324. - PubMed
-
- Matlin A.J., Clark F., Smith C.W.J. Understanding alternative splicing: towards a cellular code. Nature Rev. Mol. Cell. Biol. 2005;6:386–398. - PubMed
-
- Maniatis T., Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. - PubMed
-
- Nilsen T.W. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

