The dynamin middle domain is critical for tetramerization and higher-order self-assembly
- PMID: 17170701
- PMCID: PMC1783472
- DOI: 10.1038/sj.emboj.7601491
The dynamin middle domain is critical for tetramerization and higher-order self-assembly
Abstract
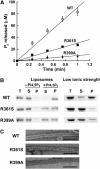
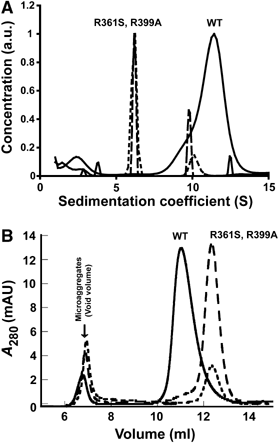
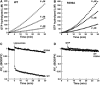
The large multidomain GTPase dynamin self-assembles around the necks of deeply invaginated coated pits at the plasma membrane and catalyzes vesicle scission by mechanisms that are not yet completely understood. Although a structural role for the 'middle' domain in dynamin function has been suggested, it has not been experimentally established. Furthermore, it is not clear whether this putative function pertains to dynamin structure in the unassembled state or to its higher-order self-assembly or both. Here, we demonstrate that two mutations in this domain, R361S and R399A, disrupt the tetrameric structure of dynamin in the unassembled state and impair its ability to stably bind to and nucleate higher-order self-assembly on membranes. Consequently, these mutations also impair dynamin's assembly-dependent stimulated GTPase activity.
Figures

References
-
- Andersson M, Wittgren B, Wahlund KG (2003) Accuracy in multiangle light scattering measurements for molar mass and radius estimations. Model calculations and experiments. Anal Chem 75: 4279–4291 - PubMed
-
- Binns DD, Barylko B, Grichine N, Atkinson MA, Helms MK, Jameson DM, Eccleston JF, Albanesi JP (1999) Correlation between self-association modes and GTPase activation of dynamin. J Protein Chem 18: 277–290 - PubMed
-
- Chen YJ, Zhang P, Egelman EH, Hinshaw JE (2004) The stalk region of dynamin drives the constriction of dynamin tubes. Nat Struct Mol Biol 11: 574–575 - PubMed
-
- Conner SD, Schmid SL (2003) Regulated portals of entry into the cell. Nature 422: 37–44 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

